Model Selection
In this notebook, the model selection capabilities of pyPESTO are demonstrated, which facilitate the selection of the best model from a set of possible models. This includes examples of forward, backward, and brute force methods, as well as criteria such as AIC, AICc, and BIC. Various additional options and convenience methods are also demonstrated.
All specification files use the PEtab Select format, which is a model selection extension to the parameter estimation specification format PEtab.
Dependencies can be installed with pip3 install pypesto[select].
In this notebook:
Example Model
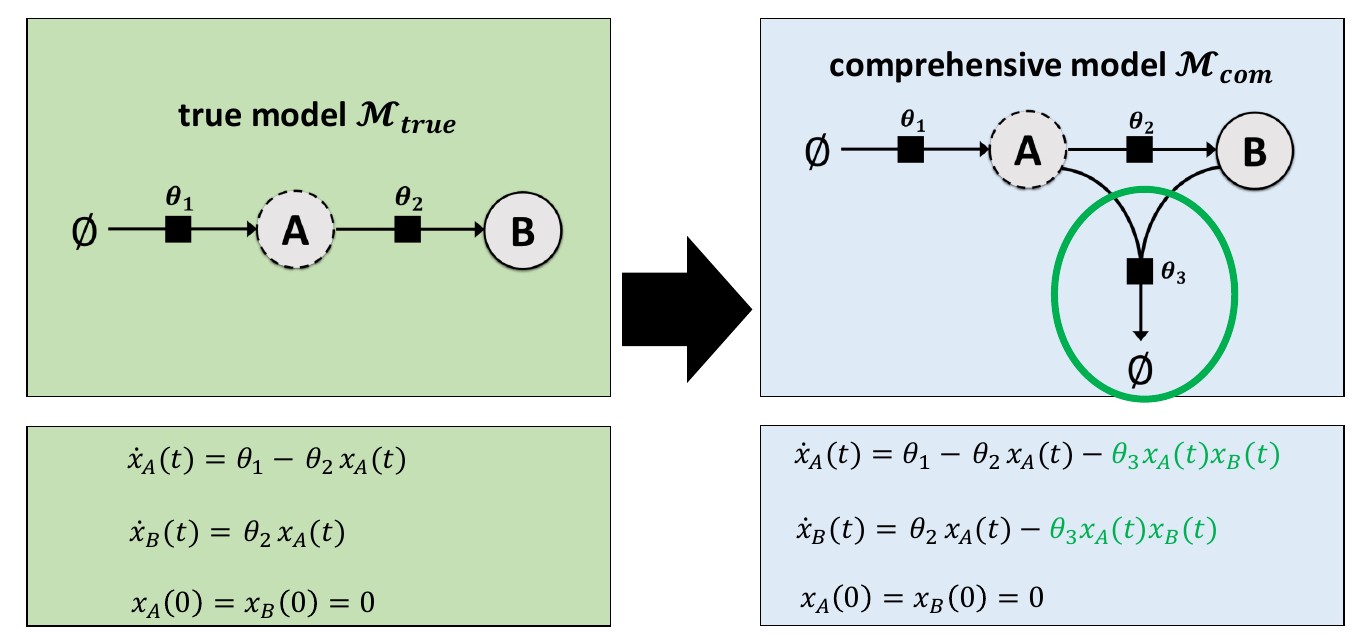
This example involves a reaction system with two species (A and B), with their growth, conversion and decay rates controlled by three parameters (\(\theta_1\), \(\theta_2\), and \(\theta_3\)). Many different hypotheses will be considered, which are described in the model specifications file. There, a parameter fixed to zero indicates that the associated reaction(s) should not be in the model which resembles a hypothesis.
Synthetic measurement data is used here, which was generated with the “true” model. The comprehensive model includes additional behavior involving a third parameter (\(\theta_3\)). Hence, during model selection, models without \(\theta_3=0\) should be preferred.
Model Space Specifications File
The model selection specification file can be written in the following compressed format.
model_subspace_id |
petab_yaml |
\(\theta_1\) |
\(\theta_2\) |
\(\theta_3\) |
|---|---|---|---|---|
M1 |
example_modelSelection.yaml |
0;estimate |
0;estimate |
0;estimate |
Alternatively, models can be explicitly specified. The below table is equivalent to the above table.
model_subspace_id |
petab_yaml |
\(\theta_1\) |
\(\theta_2\) |
\(\theta_3\) |
|---|---|---|---|---|
M1_0 |
example_modelSelection.yaml |
0 |
0 |
0 |
M1_1 |
example_modelSelection.yaml |
0 |
0 |
estimate |
M1_2 |
example_modelSelection.yaml |
0 |
estimate |
0 |
M1_3 |
example_modelSelection.yaml |
estimate |
0 |
0 |
M1_4 |
example_modelSelection.yaml |
0 |
estimate |
estimate |
M1_5 |
example_modelSelection.yaml |
estimate |
0 |
estimate |
M1_6 |
example_modelSelection.yaml |
estimate |
estimate |
0 |
M1_7 |
example_modelSelection.yaml |
estimate |
estimate |
estimate |
Either of the above tables (as TSV files) are valid inputs. Any combinations of cells in the compressed or explicit format is also acceptable, including the following example.
model_subspace_id |
petab_yaml |
\(\theta_1\) |
\(\theta_2\) |
\(\theta_3\) |
|---|---|---|---|---|
M1 |
example_modelSelection.yaml |
0;estimate |
0;estimate |
0 |
M2 |
example_modelSelection.yaml |
0;estimate |
0;estimate |
estimate |
Due to the topology of the example model, setting \(\theta_1\) to zero can result in a model with no dynamics. Hence, for this example, some parameters are set to non-zero fixed values. These parameters are considered as fixed (not estimated) values in criterion (e.g. AIC) calculations.
The model specification table used in this notebook is shown below.
[1]:
import pandas as pd
from IPython.display import HTML, display
df = pd.read_csv("model_selection/model_space.tsv", sep="\t")
display(HTML(df.to_html(index=False)))
| model_subspace_id | petab_yaml | k1 | k2 | k3 |
|---|---|---|---|---|
| M1_0 | example_modelSelection.yaml | 0 | 0 | 0 |
| M1_1 | example_modelSelection.yaml | 0.2 | 0.1 | estimate |
| M1_2 | example_modelSelection.yaml | 0.2 | estimate | 0 |
| M1_3 | example_modelSelection.yaml | estimate | 0.1 | 0 |
| M1_4 | example_modelSelection.yaml | 0.2 | estimate | estimate |
| M1_5 | example_modelSelection.yaml | estimate | 0.1 | estimate |
| M1_6 | example_modelSelection.yaml | estimate | estimate | 0 |
| M1_7 | example_modelSelection.yaml | estimate | estimate | estimate |
Forward Selection, Multiple Searches
Here, we show a typical workflow for model selection. First, a PEtab Select problem is created, which is used to initialize a pyPESTO model selection problem.
[2]:
from petab_select import VIRTUAL_INITIAL_MODEL
# Helpers for plotting etc.
def get_labels(models):
labels = {
model.get_hash(): str(model.model_subspace_id)
for model in models
if model != VIRTUAL_INITIAL_MODEL
}
return labels
def get_digraph_labels(models, criterion):
zero = min(model.get_criterion(criterion) for model in models)
labels = {
model.get_hash(): f"{model.model_subspace_id}\n{model.get_criterion(criterion) - zero:.2f}"
for model in models
}
return labels
# Disable some logged messages that make the model selection log more
# difficult to read.
import tqdm
def nop(it, *a, **k):
return it
tqdm.tqdm = nop
[3]:
import petab_select
from petab_select import ESTIMATE, Criterion, Method
import pypesto.select
petab_select_problem = petab_select.Problem.from_yaml(
"model_selection/petab_select_problem.yaml"
)
[4]:
import logging
import pypesto.logging
pypesto.logging.log(level=logging.WARNING, name="pypesto.petab", console=True)
pypesto_select_problem_1 = pypesto.select.Problem(
petab_select_problem=petab_select_problem
)
Models can be selected with a model selection algorithm (here: forward) and a comparison criterion (here: AIC). The forward method starts with the smallest model. Within each following iteration it tests all models with one additional estimated parameter.
To perform a single iteration, use select as shown below. Later in the notebook, select_to_completion is demonstrated, which performs multiple consecutive iterations automatically.
As no initial model is specified here, a virtual initial model with no estimated parameters is automatically used to find the “smallest” (in terms of number of estimated parameters) models. In this example, this is the model M1_0, which has no estimated parameters.
[5]:
# Reduce notebook runtime
minimize_options = {
"n_starts": 10,
"filename": None,
"progress_bar": False,
}
best_model_1, _ = pypesto_select_problem_1.select(
method=Method.FORWARD,
criterion=Criterion.AIC,
minimize_options=minimize_options,
)
Specifying `minimize_options` as an individual argument is deprecated. Please instead specify it within some `model_problem_options` dictionary, e.g. `model_problem_options={"minimize_options": ...}`.
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
2024-04-15 14:30:12.684 - amici.petab.sbml_import - INFO - Importing model ...
2024-04-15 14:30:12.685 - amici.petab.sbml_import - INFO - Validating PEtab problem ...
Visualization table not available. Skipping.
2024-04-15 14:30:12.697 - amici.petab.sbml_import - INFO - Model name is 'caroModel_linear'.
Writing model code to '/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear'.
2024-04-15 14:30:12.698 - amici.petab.sbml_import - INFO - Species: 2
2024-04-15 14:30:12.699 - amici.petab.sbml_import - INFO - Global parameters: 5
2024-04-15 14:30:12.699 - amici.petab.sbml_import - INFO - Reactions: 3
2024-04-15 14:30:12.732 - amici.petab.sbml_import - INFO - Observables: 1
2024-04-15 14:30:12.732 - amici.petab.sbml_import - INFO - Sigmas: 1
2024-04-15 14:30:12.735 - amici.petab.sbml_import - DEBUG - Adding output parameters to model: ['noiseParameter1_obs_x2']
2024-04-15 14:30:12.736 - amici.petab.sbml_import - DEBUG - Adding initial assignments for dict_keys([])
2024-04-15 14:30:12.739 - amici.petab.sbml_import - DEBUG - Fixed parameters are ['observable_x2']
2024-04-15 14:30:12.740 - amici.petab.sbml_import - INFO - Overall fixed parameters: 1
2024-04-15 14:30:12.741 - amici.petab.sbml_import - INFO - Variable parameters: 5
2024-04-15 14:30:12.756 - amici.sbml_import - DEBUG - Finished processing SBML annotations ++ (3.25E-04s)
2024-04-15 14:30:12.768 - amici.sbml_import - DEBUG - Finished gathering local SBML symbols ++ (5.31E-03s)
2024-04-15 14:30:12.776 - amici.sbml_import - DEBUG - Finished processing SBML parameters ++ (3.89E-04s)
2024-04-15 14:30:12.783 - amici.sbml_import - DEBUG - Finished processing SBML compartments ++ (1.17E-04s)
2024-04-15 14:30:12.797 - amici.sbml_import - DEBUG - Finished processing SBML species initials +++ (7.63E-04s)
2024-04-15 14:30:12.804 - amici.sbml_import - DEBUG - Finished processing SBML rate rules +++ (1.29E-05s)
2024-04-15 14:30:12.805 - amici.sbml_import - DEBUG - Finished processing SBML species ++ (1.56E-02s)
2024-04-15 14:30:12.814 - amici.sbml_import - DEBUG - Finished processing SBML reactions ++ (1.61E-03s)
2024-04-15 14:30:12.822 - amici.sbml_import - DEBUG - Finished processing SBML rules ++ (6.16E-04s)
2024-04-15 14:30:12.829 - amici.sbml_import - DEBUG - Finished processing SBML events ++ (4.54E-05s)
2024-04-15 14:30:12.836 - amici.sbml_import - DEBUG - Finished processing SBML initial assignments++ (2.08E-05s)
2024-04-15 14:30:12.843 - amici.sbml_import - DEBUG - Finished processing SBML species references ++ (8.38E-05s)
2024-04-15 14:30:12.844 - amici.sbml_import - DEBUG - Finished importing SBML + (9.44E-02s)
2024-04-15 14:30:12.862 - amici.sbml_import - DEBUG - Finished processing SBML observables + (1.07E-02s)
2024-04-15 14:30:12.870 - amici.sbml_import - DEBUG - Finished processing SBML event observables + (1.74E-06s)
2024-04-15 14:30:12.910 - amici.de_model - DEBUG - Finished simplifying xdot ++ (8.50E-04s)
2024-04-15 14:30:12.910 - amici.de_model - DEBUG - Finished computing xdot + (8.70E-03s)
2024-04-15 14:30:12.926 - amici.de_model - DEBUG - Finished simplifying x0 ++ (9.59E-05s)
2024-04-15 14:30:12.926 - amici.de_model - DEBUG - Finished computing x0 + (7.51E-03s)
2024-04-15 14:30:12.943 - amici.de_model - DEBUG - Finished simplifying w ++ (1.12E-03s)
2024-04-15 14:30:12.943 - amici.de_model - DEBUG - Finished computing w + (9.00E-03s)
2024-04-15 14:30:12.981 - amici.de_model - DEBUG - Finished simplifying Jy ++++ (8.24E-03s)
2024-04-15 14:30:12.981 - amici.de_model - DEBUG - Finished computing Jy +++ (1.59E-02s)
2024-04-15 14:30:13.007 - amici.de_model - DEBUG - Finished simplifying y ++++ (8.49E-05s)
2024-04-15 14:30:13.008 - amici.de_model - DEBUG - Finished computing y +++ (8.64E-03s)
2024-04-15 14:30:13.023 - amici.de_model - DEBUG - Finished simplifying sigmay ++++ (8.07E-05s)
2024-04-15 14:30:13.023 - amici.de_model - DEBUG - Finished computing sigmay +++ (7.52E-03s)
2024-04-15 14:30:13.024 - amici.de_export - DEBUG - Finished writing Jy.cpp ++ (6.47E-02s)
2024-04-15 14:30:13.066 - amici.de_model - DEBUG - Finished simplifying dJydsigma ++++ (2.92E-03s)
2024-04-15 14:30:13.066 - amici.de_model - DEBUG - Finished computing dJydsigma +++ (2.84E-02s)
2024-04-15 14:30:13.069 - amici.de_export - DEBUG - Finished writing dJydsigma.cpp ++ (3.74E-02s)
2024-04-15 14:30:13.105 - amici.de_model - DEBUG - Finished simplifying dJydy ++++ (3.74E-03s)
2024-04-15 14:30:13.106 - amici.de_model - DEBUG - Finished computing dJydy +++ (2.32E-02s)
2024-04-15 14:30:13.109 - amici.de_export - DEBUG - Finished writing dJydy.cpp ++ (3.31E-02s)
2024-04-15 14:30:13.129 - amici.de_model - DEBUG - Finished simplifying Jz ++++ (7.47E-05s)
2024-04-15 14:30:13.130 - amici.de_model - DEBUG - Finished computing Jz +++ (7.49E-03s)
2024-04-15 14:30:13.131 - amici.de_export - DEBUG - Finished writing Jz.cpp ++ (1.42E-02s)
2024-04-15 14:30:13.158 - amici.de_model - DEBUG - Finished simplifying dJzdsigma ++++ (7.13E-05s)
2024-04-15 14:30:13.159 - amici.de_model - DEBUG - Finished computing dJzdsigma +++ (1.44E-02s)
2024-04-15 14:30:13.160 - amici.de_export - DEBUG - Finished writing dJzdsigma.cpp ++ (2.12E-02s)
2024-04-15 14:30:13.187 - amici.de_model - DEBUG - Finished simplifying dJzdz ++++ (6.45E-05s)
2024-04-15 14:30:13.187 - amici.de_model - DEBUG - Finished computing dJzdz +++ (1.42E-02s)
2024-04-15 14:30:13.188 - amici.de_export - DEBUG - Finished writing dJzdz.cpp ++ (2.10E-02s)
2024-04-15 14:30:13.209 - amici.de_model - DEBUG - Finished simplifying Jrz ++++ (7.65E-05s)
2024-04-15 14:30:13.209 - amici.de_model - DEBUG - Finished computing Jrz +++ (7.59E-03s)
2024-04-15 14:30:13.210 - amici.de_export - DEBUG - Finished writing Jrz.cpp ++ (1.42E-02s)
2024-04-15 14:30:13.237 - amici.de_model - DEBUG - Finished simplifying dJrzdsigma ++++ (6.26E-05s)
2024-04-15 14:30:13.238 - amici.de_model - DEBUG - Finished computing dJrzdsigma +++ (1.41E-02s)
2024-04-15 14:30:13.239 - amici.de_export - DEBUG - Finished writing dJrzdsigma.cpp ++ (2.11E-02s)
2024-04-15 14:30:13.265 - amici.de_model - DEBUG - Finished simplifying dJrzdz ++++ (6.15E-05s)
2024-04-15 14:30:13.266 - amici.de_model - DEBUG - Finished computing dJrzdz +++ (1.42E-02s)
2024-04-15 14:30:13.267 - amici.de_export - DEBUG - Finished writing dJrzdz.cpp ++ (2.08E-02s)
2024-04-15 14:30:13.287 - amici.de_model - DEBUG - Finished simplifying root ++++ (7.05E-05s)
2024-04-15 14:30:13.288 - amici.de_model - DEBUG - Finished computing root +++ (7.29E-03s)
2024-04-15 14:30:13.289 - amici.de_export - DEBUG - Finished writing root.cpp ++ (1.42E-02s)
2024-04-15 14:30:13.318 - amici.de_model - DEBUG - Finished simplifying dwdp ++++ (4.67E-04s)
2024-04-15 14:30:13.319 - amici.de_model - DEBUG - Finished computing dwdp +++ (1.66E-02s)
2024-04-15 14:30:13.345 - amici.de_model - DEBUG - Finished simplifying dwdx ++++ (6.83E-04s)
2024-04-15 14:30:13.345 - amici.de_model - DEBUG - Finished computing dwdx +++ (1.79E-02s)
2024-04-15 14:30:13.369 - amici.de_model - DEBUG - Finished simplifying dwdw ++++ (6.86E-05s)
2024-04-15 14:30:13.369 - amici.de_model - DEBUG - Finished computing dwdw +++ (1.56E-02s)
2024-04-15 14:30:13.386 - amici.de_model - DEBUG - Finished simplifying spl ++++ (6.22E-05s)
2024-04-15 14:30:13.386 - amici.de_model - DEBUG - Finished computing spl +++ (7.62E-03s)
2024-04-15 14:30:13.401 - amici.de_model - DEBUG - Finished simplifying sspl ++++ (7.45E-05s)
2024-04-15 14:30:13.401 - amici.de_model - DEBUG - Finished computing sspl +++ (7.29E-03s)
2024-04-15 14:30:13.402 - amici.de_export - DEBUG - Finished writing dwdp.cpp ++ (1.06E-01s)
2024-04-15 14:30:13.411 - amici.de_export - DEBUG - Finished writing dwdx.cpp ++ (1.07E-03s)
2024-04-15 14:30:13.418 - amici.de_export - DEBUG - Finished writing create_splines.cpp ++ (1.26E-04s)
2024-04-15 14:30:13.445 - amici.de_model - DEBUG - Finished simplifying spline_values +++++ (6.80E-05s)
2024-04-15 14:30:13.446 - amici.de_model - DEBUG - Finished computing spline_values ++++ (8.19E-03s)
2024-04-15 14:30:13.461 - amici.de_model - DEBUG - Finished simplifying dspline_valuesdp ++++ (6.05E-05s)
2024-04-15 14:30:13.461 - amici.de_model - DEBUG - Finished computing dspline_valuesdp +++ (3.06E-02s)
2024-04-15 14:30:13.462 - amici.de_export - DEBUG - Finished writing dspline_valuesdp.cpp ++ (3.73E-02s)
2024-04-15 14:30:13.491 - amici.de_model - DEBUG - Finished simplifying spline_slopes +++++ (6.68E-05s)
2024-04-15 14:30:13.492 - amici.de_model - DEBUG - Finished computing spline_slopes ++++ (8.36E-03s)
2024-04-15 14:30:13.506 - amici.de_model - DEBUG - Finished simplifying dspline_slopesdp ++++ (5.56E-05s)
2024-04-15 14:30:13.507 - amici.de_model - DEBUG - Finished computing dspline_slopesdp +++ (3.08E-02s)
2024-04-15 14:30:13.508 - amici.de_export - DEBUG - Finished writing dspline_slopesdp.cpp ++ (3.79E-02s)
2024-04-15 14:30:13.515 - amici.de_export - DEBUG - Finished writing dwdw.cpp ++ (7.66E-06s)
2024-04-15 14:30:13.544 - amici.de_model - DEBUG - Finished simplifying dxdotdw ++++ (1.24E-04s)
2024-04-15 14:30:13.544 - amici.de_model - DEBUG - Finished computing dxdotdw +++ (1.60E-02s)
2024-04-15 14:30:13.548 - amici.de_export - DEBUG - Finished writing dxdotdw.cpp ++ (2.50E-02s)
2024-04-15 14:30:13.574 - amici.de_model - DEBUG - Finished simplifying dxdotdx_explicit ++++ (5.83E-05s)
2024-04-15 14:30:13.575 - amici.de_model - DEBUG - Finished computing dxdotdx_explicit +++ (1.39E-02s)
2024-04-15 14:30:13.576 - amici.de_export - DEBUG - Finished writing dxdotdx_explicit.cpp ++ (2.09E-02s)
2024-04-15 14:30:13.603 - amici.de_model - DEBUG - Finished simplifying dxdotdp_explicit ++++ (5.80E-05s)
2024-04-15 14:30:13.603 - amici.de_model - DEBUG - Finished computing dxdotdp_explicit +++ (1.42E-02s)
2024-04-15 14:30:13.604 - amici.de_export - DEBUG - Finished writing dxdotdp_explicit.cpp ++ (2.12E-02s)
2024-04-15 14:30:13.640 - amici.de_model - DEBUG - Finished simplifying dydx +++++ (8.63E-05s)
2024-04-15 14:30:13.641 - amici.de_model - DEBUG - Finished computing dydx ++++ (1.59E-02s)
2024-04-15 14:30:13.665 - amici.de_model - DEBUG - Finished simplifying dydw +++++ (5.74E-05s)
2024-04-15 14:30:13.666 - amici.de_model - DEBUG - Finished computing dydw ++++ (1.60E-02s)
2024-04-15 14:30:13.674 - amici.de_model - DEBUG - Finished simplifying dydx ++++ (8.51E-05s)
2024-04-15 14:30:13.675 - amici.de_model - DEBUG - Finished computing dydx +++ (5.67E-02s)
2024-04-15 14:30:13.676 - amici.de_export - DEBUG - Finished writing dydx.cpp ++ (6.42E-02s)
2024-04-15 14:30:13.712 - amici.de_model - DEBUG - Finished simplifying dydp +++++ (5.70E-05s)
2024-04-15 14:30:13.712 - amici.de_model - DEBUG - Finished computing dydp ++++ (1.60E-02s)
2024-04-15 14:30:13.721 - amici.de_model - DEBUG - Finished simplifying dydp ++++ (5.70E-05s)
2024-04-15 14:30:13.721 - amici.de_model - DEBUG - Finished computing dydp +++ (3.20E-02s)
2024-04-15 14:30:13.723 - amici.de_export - DEBUG - Finished writing dydp.cpp ++ (3.91E-02s)
2024-04-15 14:30:13.736 - amici.de_model - DEBUG - Finished computing dzdx +++ (1.22E-04s)
2024-04-15 14:30:13.736 - amici.de_export - DEBUG - Finished writing dzdx.cpp ++ (6.48E-03s)
2024-04-15 14:30:13.750 - amici.de_model - DEBUG - Finished computing dzdp +++ (9.30E-05s)
2024-04-15 14:30:13.751 - amici.de_export - DEBUG - Finished writing dzdp.cpp ++ (6.85E-03s)
2024-04-15 14:30:13.764 - amici.de_model - DEBUG - Finished computing drzdx +++ (1.09E-04s)
2024-04-15 14:30:13.764 - amici.de_export - DEBUG - Finished writing drzdx.cpp ++ (6.36E-03s)
2024-04-15 14:30:13.778 - amici.de_model - DEBUG - Finished computing drzdp +++ (8.98E-05s)
2024-04-15 14:30:13.778 - amici.de_export - DEBUG - Finished writing drzdp.cpp ++ (6.76E-03s)
2024-04-15 14:30:13.805 - amici.de_model - DEBUG - Finished simplifying dsigmaydy ++++ (6.37E-05s)
2024-04-15 14:30:13.806 - amici.de_model - DEBUG - Finished computing dsigmaydy +++ (1.44E-02s)
2024-04-15 14:30:13.807 - amici.de_export - DEBUG - Finished writing dsigmaydy.cpp ++ (2.11E-02s)
2024-04-15 14:30:13.834 - amici.de_model - DEBUG - Finished simplifying dsigmaydp ++++ (9.43E-05s)
2024-04-15 14:30:13.835 - amici.de_model - DEBUG - Finished computing dsigmaydp +++ (1.47E-02s)
2024-04-15 14:30:13.836 - amici.de_export - DEBUG - Finished writing dsigmaydp.cpp ++ (2.23E-02s)
2024-04-15 14:30:13.844 - amici.de_export - DEBUG - Finished writing sigmay.cpp ++ (3.70E-04s)
2024-04-15 14:30:13.872 - amici.de_model - DEBUG - Finished simplifying sigmaz +++++ (6.85E-05s)
2024-04-15 14:30:13.872 - amici.de_model - DEBUG - Finished computing sigmaz ++++ (8.20E-03s)
2024-04-15 14:30:13.887 - amici.de_model - DEBUG - Finished simplifying dsigmazdp ++++ (5.88E-05s)
2024-04-15 14:30:13.888 - amici.de_model - DEBUG - Finished computing dsigmazdp +++ (3.08E-02s)
2024-04-15 14:30:13.889 - amici.de_export - DEBUG - Finished writing dsigmazdp.cpp ++ (3.76E-02s)
2024-04-15 14:30:13.896 - amici.de_export - DEBUG - Finished writing sigmaz.cpp ++ (7.15E-06s)
2024-04-15 14:30:13.909 - amici.de_model - DEBUG - Finished computing stau +++ (1.13E-04s)
2024-04-15 14:30:13.910 - amici.de_export - DEBUG - Finished writing stau.cpp ++ (6.49E-03s)
2024-04-15 14:30:13.924 - amici.de_model - DEBUG - Finished computing deltax +++ (9.70E-05s)
2024-04-15 14:30:13.924 - amici.de_export - DEBUG - Finished writing deltax.cpp ++ (6.71E-03s)
2024-04-15 14:30:13.938 - amici.de_model - DEBUG - Finished computing deltasx +++ (1.38E-04s)
2024-04-15 14:30:13.939 - amici.de_export - DEBUG - Finished writing deltasx.cpp ++ (7.35E-03s)
2024-04-15 14:30:13.947 - amici.de_export - DEBUG - Finished writing w.cpp ++ (9.07E-04s)
2024-04-15 14:30:13.954 - amici.de_export - DEBUG - Finished writing x0.cpp ++ (5.49E-05s)
2024-04-15 14:30:13.975 - amici.de_model - DEBUG - Finished simplifying x0_fixedParameters ++++ (7.31E-05s)
2024-04-15 14:30:13.975 - amici.de_model - DEBUG - Finished computing x0_fixedParameters +++ (7.71E-03s)
2024-04-15 14:30:13.976 - amici.de_export - DEBUG - Finished writing x0_fixedParameters.cpp ++ (1.45E-02s)
2024-04-15 14:30:14.003 - amici.de_model - DEBUG - Finished simplifying sx0 ++++ (6.18E-05s)
2024-04-15 14:30:14.003 - amici.de_model - DEBUG - Finished computing sx0 +++ (1.34E-02s)
2024-04-15 14:30:14.005 - amici.de_export - DEBUG - Finished writing sx0.cpp ++ (2.05E-02s)
2024-04-15 14:30:14.037 - amici.de_model - DEBUG - Finished simplifying sx0_fixedParameters ++++ (5.96E-05s)
2024-04-15 14:30:14.038 - amici.de_model - DEBUG - Finished computing sx0_fixedParameters +++ (1.97E-02s)
2024-04-15 14:30:14.039 - amici.de_export - DEBUG - Finished writing sx0_fixedParameters.cpp ++ (2.64E-02s)
2024-04-15 14:30:14.047 - amici.de_export - DEBUG - Finished writing xdot.cpp ++ (1.02E-03s)
2024-04-15 14:30:14.054 - amici.de_export - DEBUG - Finished writing y.cpp ++ (2.97E-04s)
2024-04-15 14:30:14.074 - amici.de_model - DEBUG - Finished simplifying x_rdata ++++ (1.02E-04s)
2024-04-15 14:30:14.075 - amici.de_model - DEBUG - Finished computing x_rdata +++ (7.75E-03s)
2024-04-15 14:30:14.076 - amici.de_export - DEBUG - Finished writing x_rdata.cpp ++ (1.49E-02s)
2024-04-15 14:30:14.097 - amici.de_model - DEBUG - Finished simplifying total_cl ++++ (7.55E-05s)
2024-04-15 14:30:14.097 - amici.de_model - DEBUG - Finished computing total_cl +++ (7.56E-03s)
2024-04-15 14:30:14.098 - amici.de_export - DEBUG - Finished writing total_cl.cpp ++ (1.44E-02s)
2024-04-15 14:30:14.125 - amici.de_model - DEBUG - Finished simplifying dtotal_cldp ++++ (6.37E-05s)
2024-04-15 14:30:14.126 - amici.de_model - DEBUG - Finished computing dtotal_cldp +++ (1.42E-02s)
2024-04-15 14:30:14.127 - amici.de_export - DEBUG - Finished writing dtotal_cldp.cpp ++ (2.10E-02s)
2024-04-15 14:30:14.147 - amici.de_model - DEBUG - Finished simplifying dtotal_cldx_rdata ++++ (7.01E-05s)
2024-04-15 14:30:14.147 - amici.de_model - DEBUG - Finished computing dtotal_cldx_rdata +++ (7.39E-03s)
2024-04-15 14:30:14.149 - amici.de_export - DEBUG - Finished writing dtotal_cldx_rdata.cpp ++ (1.44E-02s)
2024-04-15 14:30:14.169 - amici.de_model - DEBUG - Finished simplifying x_solver ++++ (1.01E-04s)
2024-04-15 14:30:14.169 - amici.de_model - DEBUG - Finished computing x_solver +++ (7.28E-03s)
2024-04-15 14:30:14.171 - amici.de_export - DEBUG - Finished writing x_solver.cpp ++ (1.47E-02s)
2024-04-15 14:30:14.191 - amici.de_model - DEBUG - Finished simplifying dx_rdatadx_solver ++++ (1.28E-04s)
2024-04-15 14:30:14.192 - amici.de_model - DEBUG - Finished computing dx_rdatadx_solver +++ (8.01E-03s)
2024-04-15 14:30:14.193 - amici.de_export - DEBUG - Finished writing dx_rdatadx_solver.cpp ++ (1.52E-02s)
2024-04-15 14:30:14.213 - amici.de_model - DEBUG - Finished simplifying dx_rdatadp ++++ (1.50E-04s)
2024-04-15 14:30:14.214 - amici.de_model - DEBUG - Finished computing dx_rdatadp +++ (7.72E-03s)
2024-04-15 14:30:14.215 - amici.de_export - DEBUG - Finished writing dx_rdatadp.cpp ++ (1.46E-02s)
2024-04-15 14:30:14.241 - amici.de_model - DEBUG - Finished simplifying dx_rdatadtcl ++++ (6.40E-05s)
2024-04-15 14:30:14.242 - amici.de_model - DEBUG - Finished computing dx_rdatadtcl +++ (1.38E-02s)
2024-04-15 14:30:14.243 - amici.de_export - DEBUG - Finished writing dx_rdatadtcl.cpp ++ (2.08E-02s)
2024-04-15 14:30:14.257 - amici.de_model - DEBUG - Finished computing z +++ (9.09E-05s)
2024-04-15 14:30:14.258 - amici.de_export - DEBUG - Finished writing z.cpp ++ (6.71E-03s)
2024-04-15 14:30:14.271 - amici.de_model - DEBUG - Finished computing rz +++ (8.42E-05s)
2024-04-15 14:30:14.272 - amici.de_export - DEBUG - Finished writing rz.cpp ++ (6.33E-03s)
2024-04-15 14:30:14.281 - amici.de_export - DEBUG - Finished generating cpp code + (1.33E+00s)
2024-04-15 14:30:32.268 - amici.de_export - DEBUG - Finished compiling cpp code + (1.80E+01s)
2024-04-15 14:30:32.273 - amici.petab.sbml_import - INFO - Finished Importing PEtab model (1.96E+01s)
running AmiciInstall
running build_ext
------------------------------ model_ext ------------------------------
-- The C compiler identification is GNU 11.4.0
-- The CXX compiler identification is GNU 11.4.0
-- Detecting C compiler ABI info
-- Detecting C compiler ABI info - done
-- Check for working C compiler: /usr/bin/cc - skipped
-- Detecting C compile features
-- Detecting C compile features - done
-- Detecting CXX compiler ABI info
-- Detecting CXX compiler ABI info - done
-- Check for working CXX compiler: /usr/bin/c++ - skipped
-- Detecting CXX compile features
-- Detecting CXX compile features - done
-- Performing Test CUR_FLAG_SUPPORTED
-- Performing Test CUR_FLAG_SUPPORTED - Success
-- Performing Test CUR_FLAG_SUPPORTED
-- Performing Test CUR_FLAG_SUPPORTED - Success
-- Performing Test CUR_FLAG_SUPPORTED
-- Performing Test CUR_FLAG_SUPPORTED - Success
-- Performing Test CUR_FLAG_SUPPORTED
-- Performing Test CUR_FLAG_SUPPORTED - Success
-- Found OpenMP_C: -fopenmp (found version "4.5")
-- Found OpenMP_CXX: -fopenmp (found version "4.5")
-- Found OpenMP: TRUE (found version "4.5")
-- SuiteSparse_config version: 7.6.0
-- SuiteSparse_config include: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse
-- SuiteSparse_config library: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsuitesparseconfig.a
-- SuiteSparse_config static: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsuitesparseconfig.a
-- AMD version: 3.3.1
-- AMD include: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse
-- AMD library: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libamd.a
-- AMD static: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libamd.a
-- BTF version: 2.3.1
-- BTF include: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse
-- BTF library: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libbtf.a
-- BTF static: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libbtf.a
-- COLAMD version: 3.3.2
-- COLAMD include: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse
-- COLAMD library: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libcolamd.a
-- COLAMD static: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libcolamd.a
-- KLU version: 2.3.2
-- KLU include: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse
-- KLU library: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libklu.a
-- KLU static: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libklu.a
-- Looking for sgemm_
-- Looking for sgemm_ - not found
-- Performing Test CMAKE_HAVE_LIBC_PTHREAD
-- Performing Test CMAKE_HAVE_LIBC_PTHREAD - Success
-- Found Threads: TRUE
-- Looking for dgemm_
-- Looking for dgemm_ - found
-- Found BLAS: /usr/lib/x86_64-linux-gnu/libblas.so;/usr/lib/x86_64-linux-gnu/libf77blas.so;/usr/lib/x86_64-linux-gnu/libatlas.so
-- Found BLAS via FindBLAS
-- Found HDF5: /usr/lib/x86_64-linux-gnu/hdf5/serial/libhdf5.so;/usr/lib/x86_64-linux-gnu/libcrypto.so;/usr/lib/x86_64-linux-gnu/libcurl.so;/usr/lib/x86_64-linux-gnu/libpthread.a;/usr/lib/x86_64-linux-gnu/libsz.so;/usr/lib/x86_64-linux-gnu/libz.so;/usr/lib/x86_64-linux-gnu/libdl.a;/usr/lib/x86_64-linux-gnu/libm.so;/usr/lib/x86_64-linux-gnu/hdf5/serial/libhdf5_cpp.so;/usr/lib/x86_64-linux-gnu/hdf5/serial/libhdf5.so;/usr/lib/x86_64-linux-gnu/libcrypto.so;/usr/lib/x86_64-linux-gnu/libcurl.so;/usr/lib/x86_64-linux-gnu/libpthread.a;/usr/lib/x86_64-linux-gnu/libsz.so;/usr/lib/x86_64-linux-gnu/libz.so;/usr/lib/x86_64-linux-gnu/libdl.a;/usr/lib/x86_64-linux-gnu/libm.so (found version "1.10.7") found components: C HL CXX
-- Found AMICI /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/cmake/Amici
-- Could NOT find Boost (missing: Boost_INCLUDE_DIR)
-- Found SWIG: /usr/bin/swig4.0 (found version "4.0.2")
-- Found Python3: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/bin/python (found version "3.11.6") found components: Interpreter Development Development.Module Development.Embed
-- Python extension suffix is .cpython-311-x86_64-linux-gnu.so
-- Configuring done (4.6s)
-- Generating done (0.0s)
-- Build files have been written to: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext
Change Dir: '/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext'
Run Build Command(s): /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/bin/ninja -v
[1/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/caroModel_linear.cpp.o -MF CMakeFiles/caroModel_linear.dir/caroModel_linear.cpp.o.d -o CMakeFiles/caroModel_linear.dir/caroModel_linear.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/caroModel_linear.cpp
[2/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/Jy.cpp.o -MF CMakeFiles/caroModel_linear.dir/Jy.cpp.o.d -o CMakeFiles/caroModel_linear.dir/Jy.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/Jy.cpp
[3/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/create_splines.cpp.o -MF CMakeFiles/caroModel_linear.dir/create_splines.cpp.o.d -o CMakeFiles/caroModel_linear.dir/create_splines.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/create_splines.cpp
[4/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/dJydsigma.cpp.o -MF CMakeFiles/caroModel_linear.dir/dJydsigma.cpp.o.d -o CMakeFiles/caroModel_linear.dir/dJydsigma.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/dJydsigma.cpp
[5/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/dJydy.cpp.o -MF CMakeFiles/caroModel_linear.dir/dJydy.cpp.o.d -o CMakeFiles/caroModel_linear.dir/dJydy.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/dJydy.cpp
[6/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/dsigmaydp.cpp.o -MF CMakeFiles/caroModel_linear.dir/dsigmaydp.cpp.o.d -o CMakeFiles/caroModel_linear.dir/dsigmaydp.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/dsigmaydp.cpp
[7/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/dwdp.cpp.o -MF CMakeFiles/caroModel_linear.dir/dwdp.cpp.o.d -o CMakeFiles/caroModel_linear.dir/dwdp.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/dwdp.cpp
[8/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/dwdx.cpp.o -MF CMakeFiles/caroModel_linear.dir/dwdx.cpp.o.d -o CMakeFiles/caroModel_linear.dir/dwdx.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/dwdx.cpp
[9/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/dxdotdw.cpp.o -MF CMakeFiles/caroModel_linear.dir/dxdotdw.cpp.o.d -o CMakeFiles/caroModel_linear.dir/dxdotdw.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/dxdotdw.cpp
[10/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/dydx.cpp.o -MF CMakeFiles/caroModel_linear.dir/dydx.cpp.o.d -o CMakeFiles/caroModel_linear.dir/dydx.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/dydx.cpp
[11/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/sigmay.cpp.o -MF CMakeFiles/caroModel_linear.dir/sigmay.cpp.o.d -o CMakeFiles/caroModel_linear.dir/sigmay.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/sigmay.cpp
[12/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/w.cpp.o -MF CMakeFiles/caroModel_linear.dir/w.cpp.o.d -o CMakeFiles/caroModel_linear.dir/w.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/w.cpp
[13/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/x_rdata.cpp.o -MF CMakeFiles/caroModel_linear.dir/x_rdata.cpp.o.d -o CMakeFiles/caroModel_linear.dir/x_rdata.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/x_rdata.cpp
[14/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/x_solver.cpp.o -MF CMakeFiles/caroModel_linear.dir/x_solver.cpp.o.d -o CMakeFiles/caroModel_linear.dir/x_solver.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/x_solver.cpp
[15/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/y.cpp.o -MF CMakeFiles/caroModel_linear.dir/y.cpp.o.d -o CMakeFiles/caroModel_linear.dir/y.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/y.cpp
[16/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/xdot.cpp.o -MF CMakeFiles/caroModel_linear.dir/xdot.cpp.o.d -o CMakeFiles/caroModel_linear.dir/xdot.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/xdot.cpp
[17/21] /usr/bin/c++ -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT CMakeFiles/caroModel_linear.dir/wrapfunctions.cpp.o -MF CMakeFiles/caroModel_linear.dir/wrapfunctions.cpp.o.d -o CMakeFiles/caroModel_linear.dir/wrapfunctions.cpp.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/wrapfunctions.cpp
[18/21] : && /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/cmake/data/bin/cmake -E rm -f libcaroModel_linear.a && /usr/bin/ar qc libcaroModel_linear.a CMakeFiles/caroModel_linear.dir/Jy.cpp.o CMakeFiles/caroModel_linear.dir/caroModel_linear.cpp.o CMakeFiles/caroModel_linear.dir/create_splines.cpp.o CMakeFiles/caroModel_linear.dir/dJydsigma.cpp.o CMakeFiles/caroModel_linear.dir/dJydy.cpp.o CMakeFiles/caroModel_linear.dir/dsigmaydp.cpp.o CMakeFiles/caroModel_linear.dir/dwdp.cpp.o CMakeFiles/caroModel_linear.dir/dwdx.cpp.o CMakeFiles/caroModel_linear.dir/dxdotdw.cpp.o CMakeFiles/caroModel_linear.dir/dydx.cpp.o CMakeFiles/caroModel_linear.dir/sigmay.cpp.o CMakeFiles/caroModel_linear.dir/w.cpp.o CMakeFiles/caroModel_linear.dir/wrapfunctions.cpp.o CMakeFiles/caroModel_linear.dir/x_rdata.cpp.o CMakeFiles/caroModel_linear.dir/x_solver.cpp.o CMakeFiles/caroModel_linear.dir/xdot.cpp.o CMakeFiles/caroModel_linear.dir/y.cpp.o && /usr/bin/ranlib libcaroModel_linear.a && :
[19/21] cd /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext/swig && /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/cmake/data/bin/cmake -E make_directory /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext/swig/CMakeFiles/_caroModel_linear.dir /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext/swig /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext/swig/CMakeFiles/_caroModel_linear.dir && /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/cmake/data/bin/cmake -E env SWIG_LIB=/usr/share/swig4.0 /usr/bin/swig4.0 -python -I/home/docs/.asdf/installs/python/3.11.6/include/python3.11 -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/swig/.. -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/../swig -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -I/usr/include/hdf5/serial -outdir /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext/swig -c++ -interface _caroModel_linear -I/home/docs/.asdf/installs/python/3.11.6/include/python3.11 -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/swig/.. -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/../swig -o /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext/swig/CMakeFiles/_caroModel_linear.dir/caroModel_linearPYTHON_wrap.cxx /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/swig/caroModel_linear.i
[20/21] /usr/bin/c++ -D_caroModel_linear_EXPORTS -I/home/docs/.asdf/installs/python/3.11.6/include/python3.11 -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/swig/.. -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/../swig -I/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/share/amici/swig -isystem /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/include/suitesparse -isystem /usr/include/hdf5/serial -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -std=gnu++17 -fPIC -fopenmp -MD -MT swig/CMakeFiles/_caroModel_linear.dir/CMakeFiles/_caroModel_linear.dir/caroModel_linearPYTHON_wrap.cxx.o -MF swig/CMakeFiles/_caroModel_linear.dir/CMakeFiles/_caroModel_linear.dir/caroModel_linearPYTHON_wrap.cxx.o.d -o swig/CMakeFiles/_caroModel_linear.dir/CMakeFiles/_caroModel_linear.dir/caroModel_linearPYTHON_wrap.cxx.o -c /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext/swig/CMakeFiles/_caroModel_linear.dir/caroModel_linearPYTHON_wrap.cxx
[21/21] : && /usr/bin/c++ -fPIC -Wall -Wno-unused-function -Wno-unused-variable -Wno-unused-but-set-variable -O3 -DNDEBUG -shared -o swig/_caroModel_linear.cpython-311-x86_64-linux-gnu.so swig/CMakeFiles/_caroModel_linear.dir/CMakeFiles/_caroModel_linear.dir/caroModel_linearPYTHON_wrap.cxx.o -Wl,-rpath,/home/docs/.asdf/installs/python/3.11.6/lib:/usr/lib/x86_64-linux-gnu/hdf5/serial /home/docs/.asdf/installs/python/3.11.6/lib/libpython3.11.so libcaroModel_linear.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libamici.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_generic.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_nvecserial.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsolband.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunmatrixband.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsoldense.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunmatrixdense.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsolpcg.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsolspbcgs.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsolspfgmr.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsolspgmr.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsolsptfqmr.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunlinsolklu.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunmatrixsparse.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libklu.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libcolamd.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libbtf.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libamd.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsuitesparseconfig.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunnonlinsolnewton.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_sunnonlinsolfixedpoint.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_cvodes.a /home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/libsundials_idas.a -lm /usr/lib/gcc/x86_64-linux-gnu/11/libgomp.so /usr/lib/x86_64-linux-gnu/libpthread.a -ldl /usr/lib/x86_64-linux-gnu/libblas.so /usr/lib/x86_64-linux-gnu/libf77blas.so /usr/lib/x86_64-linux-gnu/libatlas.so /usr/lib/x86_64-linux-gnu/hdf5/serial/libhdf5_hl_cpp.so /usr/lib/x86_64-linux-gnu/hdf5/serial/libhdf5_hl.so /usr/lib/x86_64-linux-gnu/hdf5/serial/libhdf5_cpp.so /usr/lib/x86_64-linux-gnu/hdf5/serial/libhdf5.so && :
-- Install configuration: "Release"
-- Installing: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/caroModel_linear/./caroModel_linear.py
-- Installing: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/caroModel_linear/./_caroModel_linear.cpython-311-x86_64-linux-gnu.so
-- Installing: /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/caroModel_linear/lib/libcaroModel_linear.a
------------------------------ model_ext ------------------------------
running AmiciBuildCMakeExtension
==> Configuring:
$ cmake -S /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear -B /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext -GNinja -DCMAKE_BUILD_TYPE=Release -DCMAKE_INSTALL_PREFIX:PATH=/home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/caroModel_linear -DCMAKE_MAKE_PROGRAM=/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/bin/ninja -DCMAKE_VERBOSE_MAKEFILE=ON -DCMAKE_MODULE_PATH=/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib/cmake/SuiteSparse;/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici/lib64/cmake/SuiteSparse -DKLU_ROOT=/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/amici -DAMICI_PYTHON_BUILD_EXT_ONLY=ON -DPython3_EXECUTABLE=/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/bin/python
==> Building:
$ cmake --build /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext --config Release
==> Installing:
$ cmake --install /home/docs/checkouts/readthedocs.org/user_builds/pypesto/checkouts/stable/doc/example/amici_models/0.23.1/caroModel_linear/build_model_ext
virtual_init | M1_0:649bcd9 | AIC | None | 3.698e+01 | None | True
To search more of the model space, hence models with more parameters, the algorithm can be repeated. As models with no estimated parameters have already been tested, subsequent select calls will begin with the next simplest model (in this case, models with exactly 1 estimated parameter, if they exist in the model space), and move on to more complex models.
The best model from the first iteration is supplied as the predecessor (initial) model here.
[6]:
best_model_2, _ = pypesto_select_problem_1.select(
method=Method.FORWARD,
criterion=Criterion.AIC,
minimize_options=minimize_options,
predecessor_model=best_model_1,
)
Specifying `minimize_options` as an individual argument is deprecated. Please instead specify it within some `model_problem_options` dictionary, e.g. `model_problem_options={"minimize_options": ...}`.
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
M1_0:649bcd9 | M1_1:9202ace | AIC | 3.698e+01 | -4.175e+00 | -4.115e+01 | True
Visualization table not available. Skipping.
M1_0:649bcd9 | M1_2:c773362 | AIC | 3.698e+01 | -4.275e+00 | -4.125e+01 | True
Visualization table not available. Skipping.
M1_0:649bcd9 | M1_3:e03f5dc | AIC | 3.698e+01 | -4.705e+00 | -4.168e+01 | True
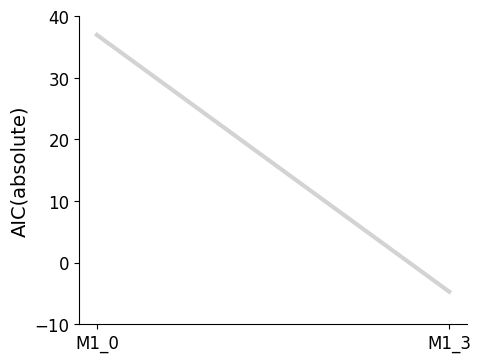
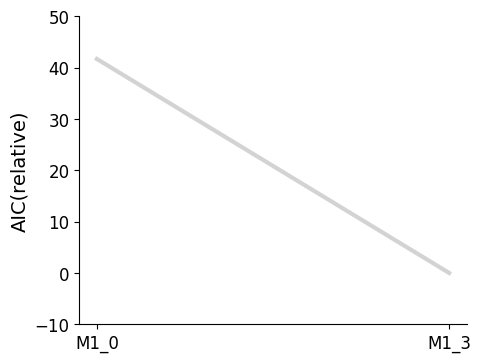
Plotting routines to visualize the best model at each iteration of the selection process, or to visualize the graph of models that have been visited in the model space are available.
[7]:
import pypesto.visualize.select as pvs
selected_models = [best_model_1, best_model_2]
ax = pvs.plot_selected_models(
[best_model_1, best_model_2],
criterion=Criterion.AIC,
relative=False,
labels=get_labels(selected_models),
)
ax = pvs.plot_selected_models(
[best_model_1, best_model_2],
criterion=Criterion.AIC,
labels=get_labels(selected_models),
)
ax.plot();
[8]:
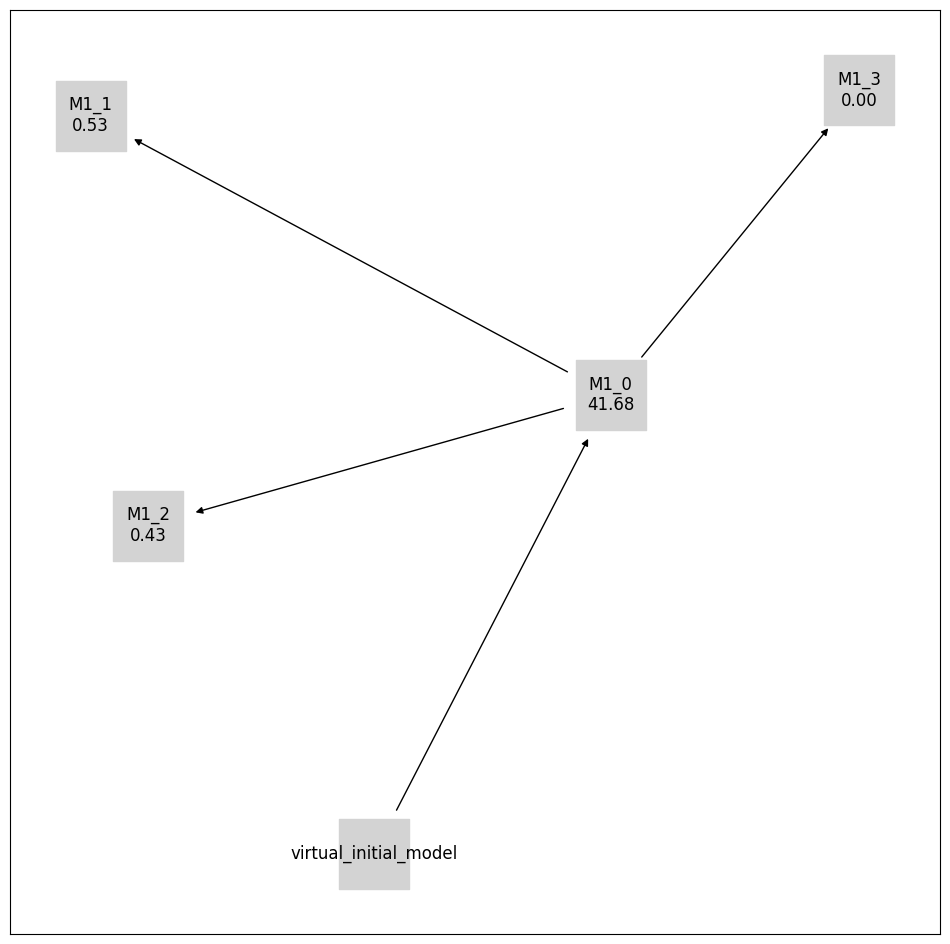
pvs.plot_calibrated_models_digraph(
problem=pypesto_select_problem_1,
labels=get_digraph_labels(
pypesto_select_problem_1.calibrated_models.values(),
criterion=Criterion.AIC,
),
);
Backward Selection, Custom Initial Model
Backward selection is specified by changing the algorithm from Method.FORWARD to Method.BACKWARD in the select() call.
A custom initial model is specified with the optional predecessor_model argument of select().
[9]:
import numpy as np
from petab_select import Model
petab_select_problem.model_space.reset_exclusions()
pypesto_select_problem_2 = pypesto.select.Problem(
petab_select_problem=petab_select_problem
)
petab_yaml = "model_selection/example_modelSelection.yaml"
initial_model = Model(
model_id="myModel",
petab_yaml=petab_yaml,
parameters={
"k1": 0.1,
"k2": ESTIMATE,
"k3": ESTIMATE,
},
criteria={petab_select_problem.criterion: np.inf},
)
print("Initial model:")
print(initial_model)
Initial model:
model_id petab_yaml k1 k2 k3
myModel model_selection/example_modelSelection.yaml 0.1 estimate estimate
[10]:
pypesto_select_problem_2.select(
method=Method.BACKWARD,
criterion=Criterion.AIC,
predecessor_model=initial_model,
minimize_options=minimize_options,
);
Specifying `minimize_options` as an individual argument is deprecated. Please instead specify it within some `model_problem_options` dictionary, e.g. `model_problem_options={"minimize_options": ...}`.
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
:myModel | M1_1:9202ace | AIC | inf | 3.897e+01 | -inf | True
Visualization table not available. Skipping.
:myModel | M1_2:c773362 | AIC | inf | -4.275e+00 | -inf | True
[11]:
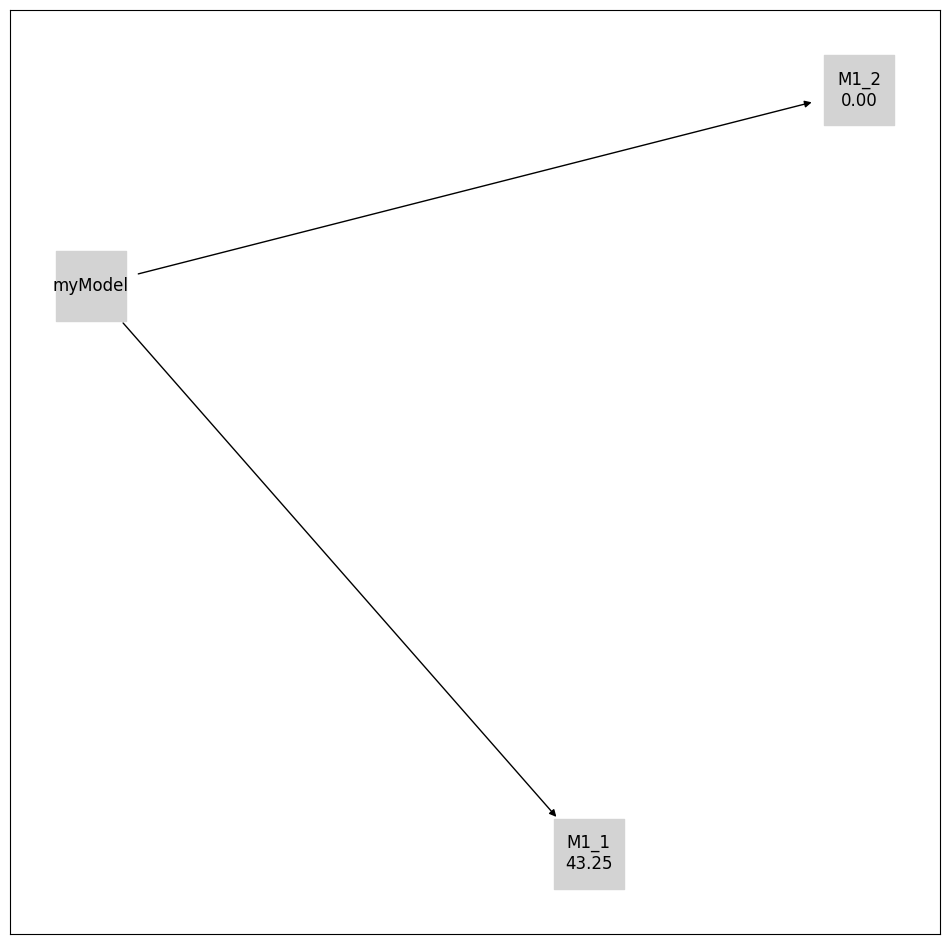
initial_model_label = {initial_model.get_hash(): initial_model.model_id}
pvs.plot_calibrated_models_digraph(
problem=pypesto_select_problem_2,
labels={
**get_digraph_labels(
pypesto_select_problem_2.calibrated_models.values(),
criterion=Criterion.AIC,
),
**initial_model_label,
},
);
Additional Options
There exist additional options that can be used to further customise selection algorithms.
Select First Improvement
At each selection step, as soon as a model that improves on the previous model is encountered (by the specified criterion), it is selected and immediately used as the previous model in the next iteration of the selection. This is unlike the default behaviour, where all test models at each iteration are optimized, and the best of these is selected.
Use Previous Maximum Likelihood Estimate as Startpoint
The maximum likelihood estimate from the previous model is used as one of the startpoints in the multistart optimization of the test models. The default behaviour is that all startpoints are automatically generated by pyPESTO.
Minimize Options
Optimization can be customised with a dictionary that specifies values for the corresponding keyword arguments of minimize.
Criterion Options
Currently implemented options are: Criterion.AIC (Akaike information criterion), Criterion.AICC (corrected AIC), and Criterion.BIC (Bayesian information criterion).
Criterion Threshold
A threshold can be specified, such that only models that improve on previous models by the threshold amount in the chosen criterion are accepted.
[12]:
petab_select_problem.model_space.reset_exclusions()
pypesto_select_problem_3 = pypesto.select.Problem(
petab_select_problem=petab_select_problem
)
best_models = pypesto_select_problem_3.select_to_completion(
method=Method.FORWARD,
criterion=Criterion.BIC,
select_first_improvement=True,
startpoint_latest_mle=True,
minimize_options=minimize_options,
)
Specifying `minimize_options` as an individual argument is deprecated. Please instead specify it within some `model_problem_options` dictionary, e.g. `model_problem_options={"minimize_options": ...}`.
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
virtual_init | M1_0:649bcd9 | BIC | None | 3.677e+01 | None | True
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
M1_0:649bcd9 | M1_1:9202ace | BIC | 3.677e+01 | -4.592e+00 | -4.136e+01 | True
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
M1_1:9202ace | M1_4:3d668e2 | BIC | -4.592e+00 | -2.899e+00 | 1.693e+00 | False
Visualization table not available. Skipping.
M1_1:9202ace | M1_5:d2503eb | BIC | -4.592e+00 | -3.330e+00 | 1.262e+00 | False
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
M1_1:9202ace | M1_7:afa580e | BIC | -4.592e+00 | -4.889e+00 | -2.975e-01 | True
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
[13]:
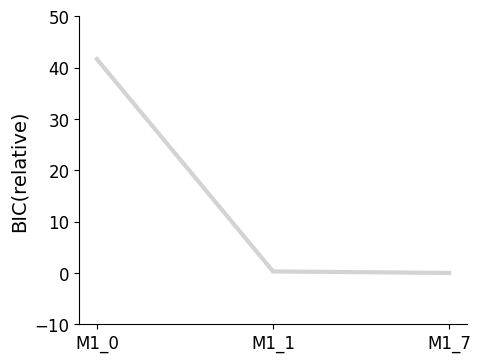
pvs.plot_selected_models(
selected_models=best_models,
criterion=Criterion.BIC,
labels=get_labels(best_models),
);
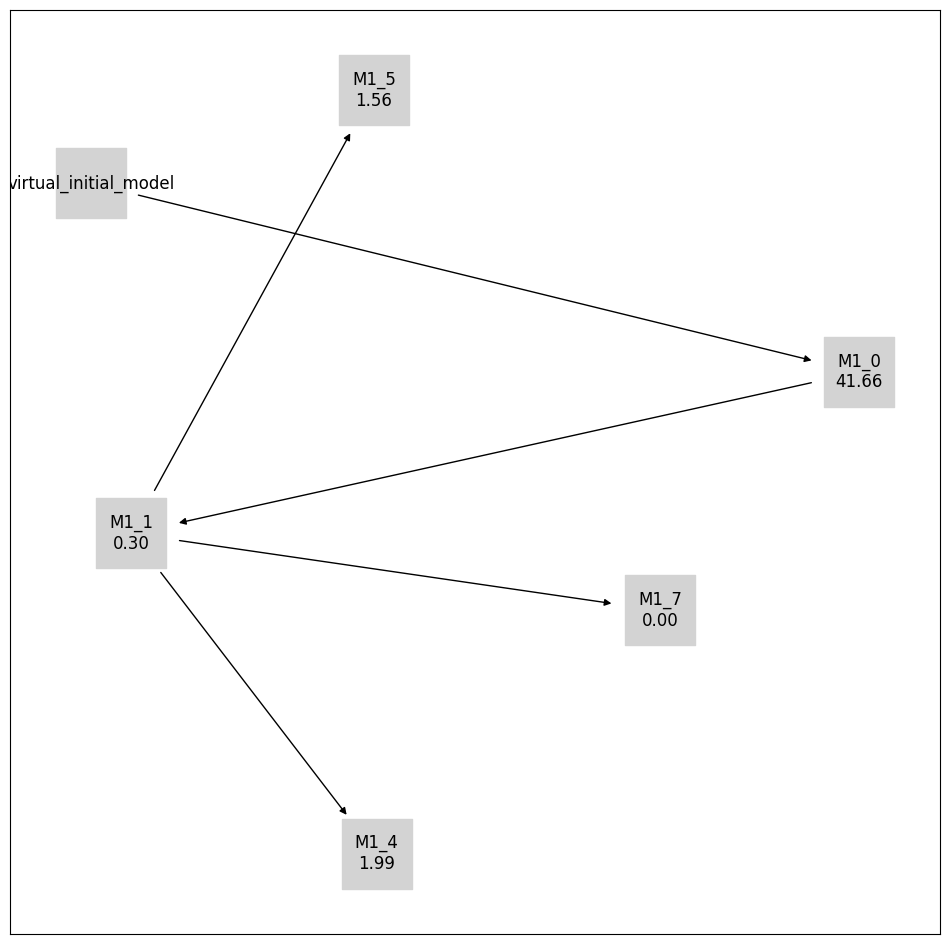
[14]:
pvs.plot_calibrated_models_digraph(
problem=pypesto_select_problem_3,
criterion=Criterion.BIC,
relative=False,
labels=get_digraph_labels(
pypesto_select_problem_3.calibrated_models.values(),
criterion=Criterion.BIC,
),
);
[15]:
# Repeat with AICc and criterion_threshold == 10
petab_select_problem.model_space.reset_exclusions()
pypesto_select_problem_4 = pypesto.select.Problem(
petab_select_problem=petab_select_problem
)
best_models = pypesto_select_problem_4.select_to_completion(
method=Method.FORWARD,
criterion=Criterion.AICC,
select_first_improvement=True,
startpoint_latest_mle=True,
minimize_options=minimize_options,
criterion_threshold=10,
)
Specifying `minimize_options` as an individual argument is deprecated. Please instead specify it within some `model_problem_options` dictionary, e.g. `model_problem_options={"minimize_options": ...}`.
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
virtual_init | M1_0:649bcd9 | AICc | None | 3.798e+01 | None | True
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
M1_0:649bcd9 | M1_1:9202ace | AICc | 3.798e+01 | -1.754e-01 | -3.815e+01 | True
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
M1_1:9202ace | M1_4:3d668e2 | AICc | -1.754e-01 | 9.725e+00 | 9.901e+00 | False
Visualization table not available. Skipping.
M1_1:9202ace | M1_5:d2503eb | AICc | -1.754e-01 | 9.295e+00 | 9.470e+00 | False
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
M1_1:9202ace | M1_7:afa580e | AICc | -1.754e-01 | 3.594e+01 | 3.612e+01 | False
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
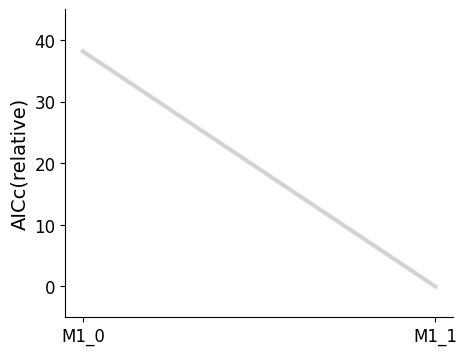
[16]:
pvs.plot_selected_models(
selected_models=best_models,
criterion=Criterion.AICC,
labels=get_labels(best_models),
);
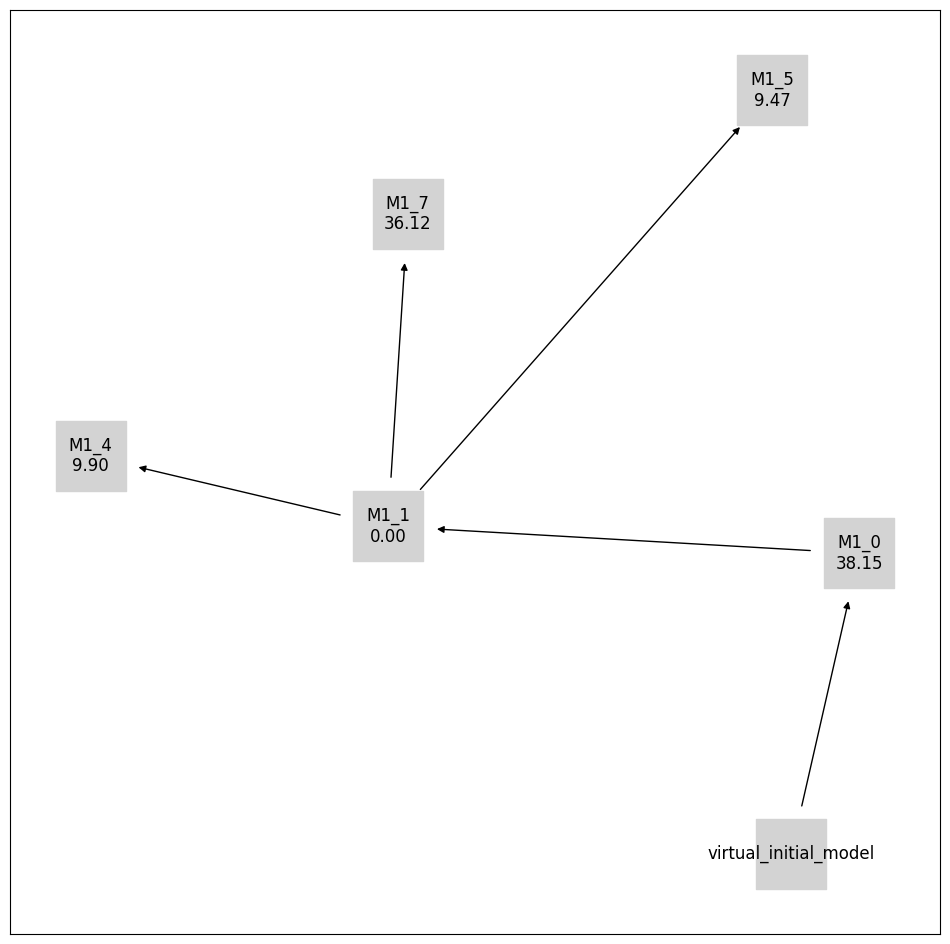
[17]:
pvs.plot_calibrated_models_digraph(
problem=pypesto_select_problem_4,
criterion=Criterion.AICC,
relative=False,
labels=get_digraph_labels(
pypesto_select_problem_4.calibrated_models.values(),
criterion=Criterion.AICC,
),
);
Multistart
Multiple model selections can be run by specifying multiple initial models.
[18]:
petab_select_problem.model_space.reset_exclusions()
pypesto_select_problem_5 = pypesto.select.Problem(
petab_select_problem=petab_select_problem
)
initial_model_1 = Model(
model_id="myModel1",
petab_yaml=petab_yaml,
parameters={
"k1": 0,
"k2": 0,
"k3": 0,
},
criteria={petab_select_problem.criterion: np.inf},
)
initial_model_2 = Model(
model_id="myModel2",
petab_yaml=petab_yaml,
parameters={
"k1": ESTIMATE,
"k2": ESTIMATE,
"k3": 0,
},
criteria={petab_select_problem.criterion: np.inf},
)
initial_models = [initial_model_1, initial_model_2]
best_model, best_models = pypesto_select_problem_5.multistart_select(
method=Method.FORWARD,
criterion=Criterion.AIC,
predecessor_models=initial_models,
minimize_options=minimize_options,
)
Specifying `minimize_options` as an individual argument is deprecated. Please instead specify it within some `model_problem_options` dictionary, e.g. `model_problem_options={"minimize_options": ...}`.
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
:myModel1 | M1_1:9202ace | AIC | inf | 3.897e+01 | -inf | True
Visualization table not available. Skipping.
:myModel1 | M1_2:c773362 | AIC | inf | 2.412e+01 | -inf | True
Visualization table not available. Skipping.
:myModel1 | M1_3:e03f5dc | AIC | inf | -4.705e+00 | -inf | True
--------------------New Selection--------------------
model0 | model | crit | model0_crit | model_crit | crit_diff | accept
Visualization table not available. Skipping.
:myModel2 | M1_7:afa580e | AIC | inf | 1.934e+01 | -inf | True
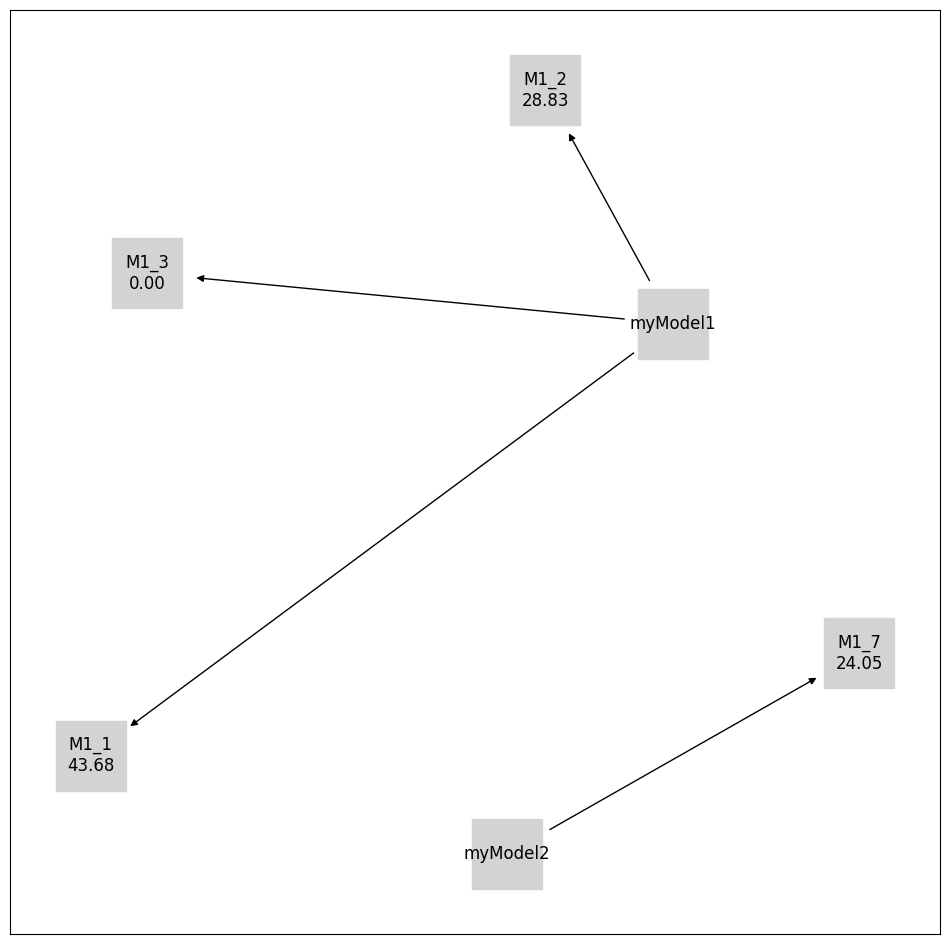
[19]:
initial_model_labels = {
initial_model.get_hash(): initial_model.model_id
for initial_model in initial_models
}
pvs.plot_calibrated_models_digraph(
problem=pypesto_select_problem_5,
criterion=Criterion.AIC,
relative=False,
labels={
**get_digraph_labels(
pypesto_select_problem_5.calibrated_models.values(),
criterion=Criterion.AIC,
),
**initial_model_labels,
},
);