Conversion reaction
[1]:
# install if not done yet
# !apt install libatlas-base-dev swig
# %pip install pypesto[amici] --quiet
[2]:
import importlib
import os
import sys
import amici
import amici.plotting
import numpy as np
import pypesto
import pypesto.optimize as optimize
import pypesto.visualize as visualize
# sbml file we want to import
sbml_file = "conversion_reaction/model_conversion_reaction.xml"
# name of the model that will also be the name of the python module
model_name = "model_conversion_reaction"
# directory to which the generated model code is written
model_output_dir = "tmp/" + model_name
Compile AMICI model
[3]:
# import sbml model, compile and generate amici module
sbml_importer = amici.SbmlImporter(sbml_file)
sbml_importer.sbml2amici(model_name, model_output_dir, verbose=False)
Load AMICI model
[4]:
# load amici module (the usual starting point later for the analysis)
sys.path.insert(0, os.path.abspath(model_output_dir))
model_module = importlib.import_module(model_name)
model = model_module.getModel()
model.requireSensitivitiesForAllParameters()
model.setTimepoints(np.linspace(0, 10, 11))
model.setParameterScale(amici.ParameterScaling.log10)
model.setParameters([-0.3, -0.7])
solver = model.getSolver()
solver.setSensitivityMethod(amici.SensitivityMethod.forward)
solver.setSensitivityOrder(amici.SensitivityOrder.first)
# how to run amici now:
rdata = amici.runAmiciSimulation(model, solver, None)
amici.plotting.plotStateTrajectories(rdata)
edata = amici.ExpData(rdata, 0.2, 0.0)
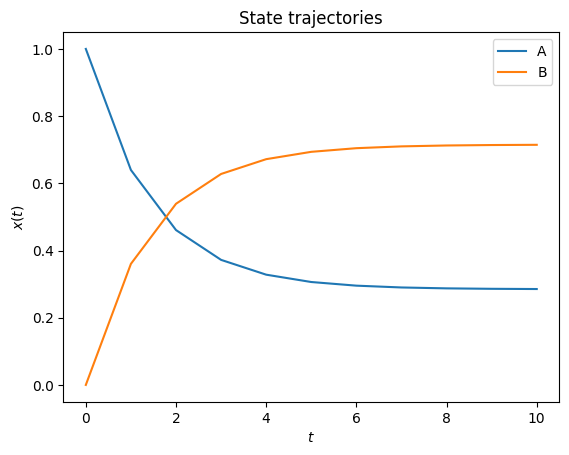
Optimize
[5]:
# create objective function from amici model
# pesto.AmiciObjective is derived from pesto.Objective,
# the general pesto objective function class
objective = pypesto.AmiciObjective(model, solver, [edata], 1)
# create optimizer object which contains all information for doing the optimization
optimizer = optimize.ScipyOptimizer(method="ls_trf")
# create problem object containing all information on the problem to be solved
problem = pypesto.Problem(objective=objective, lb=[-2, -2], ub=[2, 2])
# do the optimization
result = optimize.minimize(
problem=problem, optimizer=optimizer, n_starts=100, filename=None
)
Visualize
[6]:
visualize.waterfall(result)
visualize.parameters(result)
visualize.optimization_scatter(result=result)
[6]:
<seaborn.axisgrid.PairGrid at 0x7fb769593010>
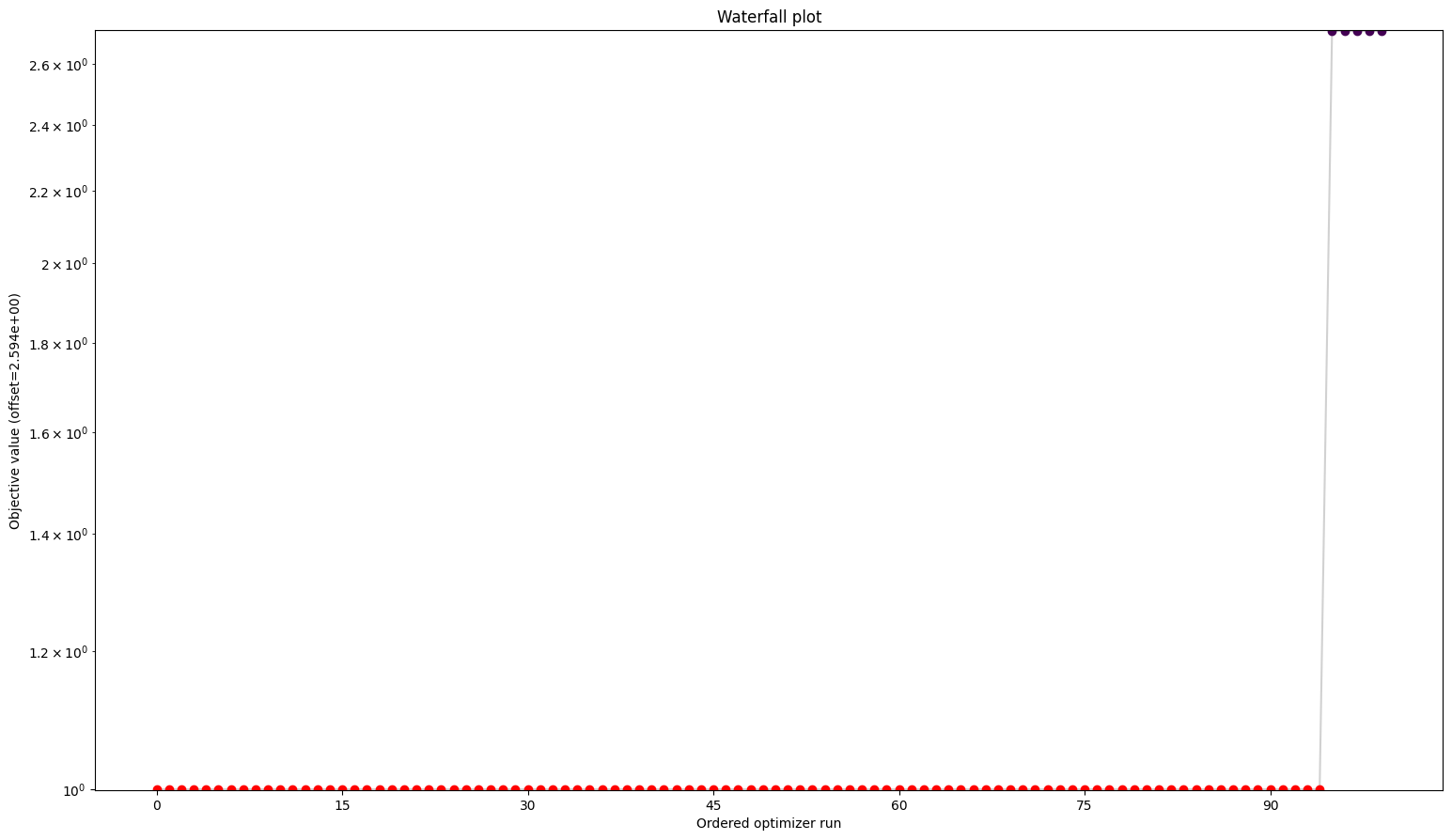
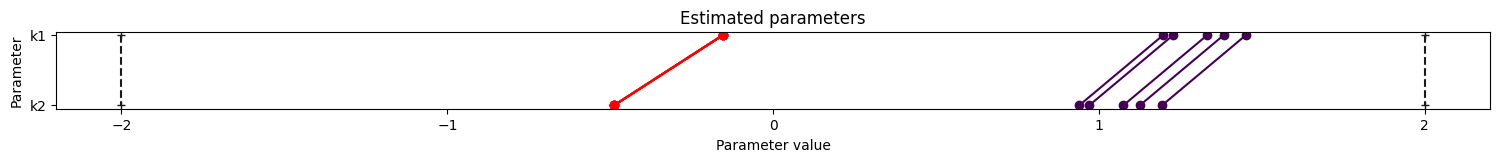
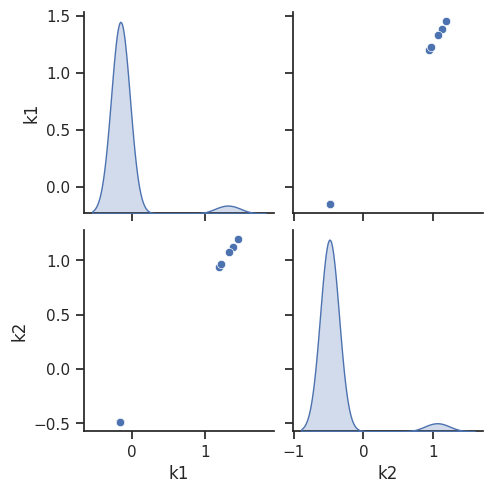
Profiles
[7]:
import pypesto.profile as profile
profile_options = profile.ProfileOptions(
min_step_size=0.0005,
delta_ratio_max=0.05,
default_step_size=0.005,
ratio_min=0.01,
)
result = profile.parameter_profile(
problem=problem,
result=result,
optimizer=optimizer,
profile_index=np.array([1, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0]),
result_index=0,
profile_options=profile_options,
filename=None,
)
[8]:
# specify the parameters, for which profiles should be computed
ax = visualize.profiles(result)
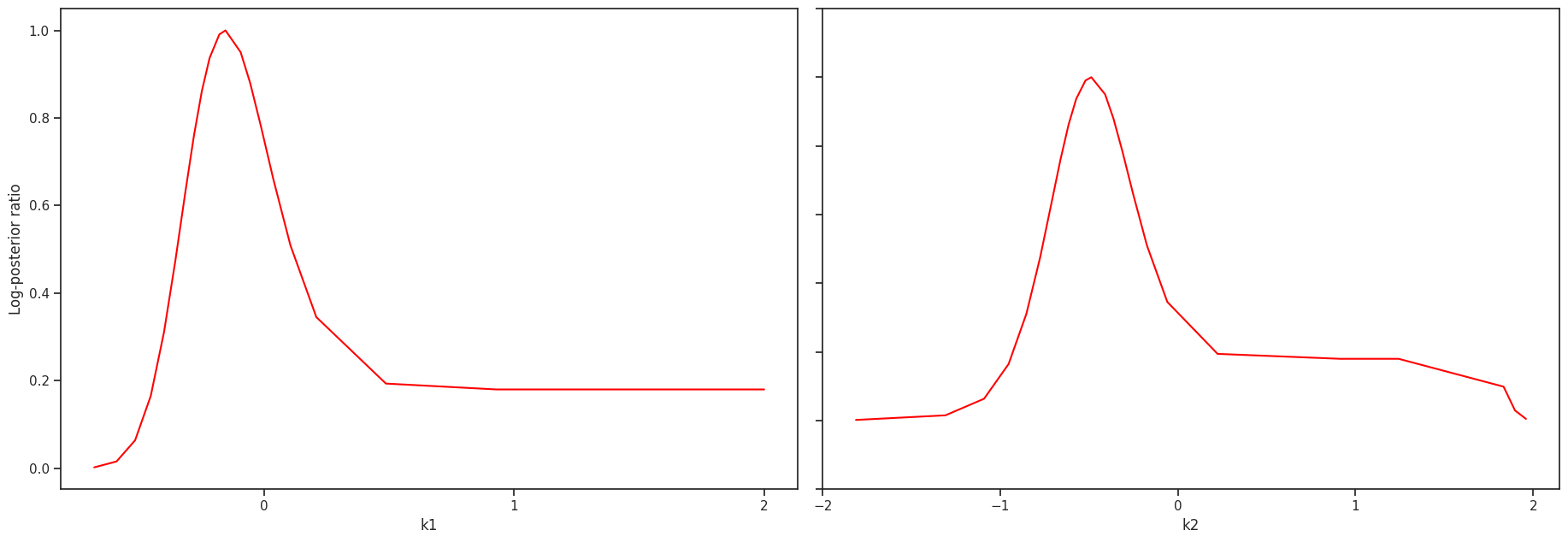
Sampling
[9]:
import pypesto.sample as sample
sampler = sample.AdaptiveParallelTemperingSampler(
internal_sampler=sample.AdaptiveMetropolisSampler(), n_chains=3
)
result = sample.sample(
problem, n_samples=10000, sampler=sampler, result=result, filename=None
)
Initializing betas with "near-exponential decay".
Elapsed time: 58.945134832
[10]:
ax = visualize.sampling_scatter(result, size=[13, 6])
/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/stable/lib/python3.11/site-packages/pypesto/visualize/sampling.py:1223: UserWarning: Burn in index not found in the results, the full chain will be shown.
You may want to use, e.g., `pypesto.sample.geweke_test`.
nr_params, params_fval, theta_lb, theta_ub, _ = get_data_to_plot(
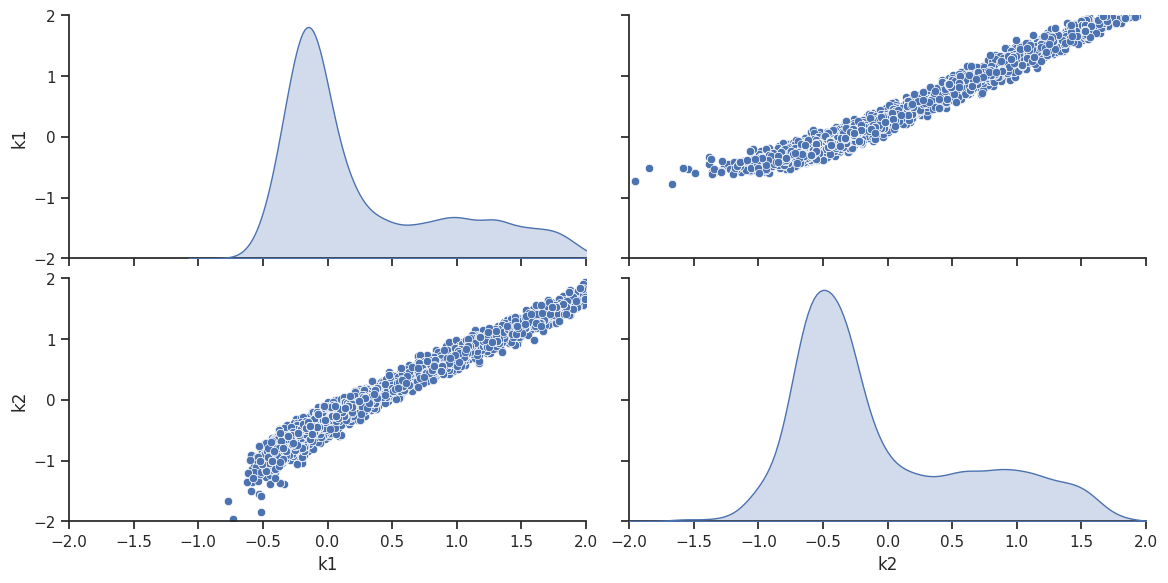
Predict
[11]:
# Let's create a function, which predicts the ratio of x_1 and x_0
import pypesto.predict as predict
def ratio_function(amici_output_list):
# This (optional) function post-processes the results from AMICI and must accept one input:
# a list of dicts, with the fields t, x, y[, sx, sy - if sensi_orders includes 1]
# We need to specify the simulation condition: here, we only have one, i.e., it's [0]
x = amici_output_list[0]["x"]
ratio = x[:, 1] / x[:, 0]
# we need to output also at least one simulation condition
return [ratio]
# create pypesto prediction function
predictor = predict.AmiciPredictor(
objective, post_processor=ratio_function, output_ids=["ratio"]
)
# run prediction
prediction = predictor(x=model.getUnscaledParameters())
[12]:
dict(prediction)
[12]:
{'conditions': [{'timepoints': array([ 0., 1., 2., 3., 4., 5., 6., 7., 8., 9., 10.]),
'output_ids': ['ratio'],
'x_names': ['k1', 'k2'],
'output': array([0. , 1.95196396, 2.00246152, 2.00290412, 2.00290796,
2.00290801, 2.00290801, 2.00290799, 2.002908 , 2.00290801,
2.002908 ]),
'output_sensi': None,
'output_weight': None,
'output_sigmay': None}],
'condition_ids': ['condition_0'],
'comment': None,
'parameter_ids': ['k1', 'k2']}
Analyze parameter ensembles
[13]:
# We want to use the sample result to create a prediction
from pypesto.ensemble import ensemble
# first collect some vectors from the sampling result
vectors = result.sample_result.trace_x[0, ::20, :]
# create the collection
my_ensemble = ensemble.Ensemble(
vectors,
x_names=problem.x_names,
ensemble_type=ensemble.EnsembleType.sample,
lower_bound=problem.lb,
upper_bound=problem.ub,
)
# we can also perform an approximative identifiability analysis
summary = my_ensemble.compute_summary()
identifiability = my_ensemble.check_identifiability()
print(identifiability.transpose())
parameterId k1 k2
parameterId k1 k2
lowerBound -2 -2
upperBound 2 2
ensemble_mean -0.321071 -0.24397
ensemble_std 0.166502 0.163219
ensemble_median -0.321071 -0.24397
within lb: 1 std True True
within ub: 1 std True True
within lb: 2 std True True
within ub: 2 std True True
within lb: 3 std True True
within ub: 3 std True True
within lb: perc 5 True True
within lb: perc 20 True True
within ub: perc 80 True True
within ub: perc 95 True True
[14]:
# run a prediction
ensemble_prediction = my_ensemble.predict(
predictor, prediction_id="ratio_over_time"
)
# go for some analysis
prediction_summary = ensemble_prediction.compute_summary(
percentiles_list=(1, 5, 10, 25, 75, 90, 95, 99)
)
dict(prediction_summary)
[14]:
{'mean': <pypesto.result.predict.PredictionResult at 0x7fb76828f310>,
'std': <pypesto.result.predict.PredictionResult at 0x7fb76828ead0>,
'median': <pypesto.result.predict.PredictionResult at 0x7fb76828d2d0>,
'percentile 1': <pypesto.result.predict.PredictionResult at 0x7fb76828edd0>,
'percentile 5': <pypesto.result.predict.PredictionResult at 0x7fb76828f290>,
'percentile 10': <pypesto.result.predict.PredictionResult at 0x7fb76828ef50>,
'percentile 25': <pypesto.result.predict.PredictionResult at 0x7fb76828d290>,
'percentile 75': <pypesto.result.predict.PredictionResult at 0x7fb76828e850>,
'percentile 90': <pypesto.result.predict.PredictionResult at 0x7fb76828c410>,
'percentile 95': <pypesto.result.predict.PredictionResult at 0x7fb76828ec10>,
'percentile 99': <pypesto.result.predict.PredictionResult at 0x7fb76828d050>}