pyPESTO: Getting started
This notebook takes you through the first steps to get started with pyPESTO.

pyPESTO is a python package for parameter inference, offering a unified interface to various optimization and sampling methods. pyPESTO is highly modular and customizable, e.g., with respect to objective function definition and employed inference algorithms.
[1]:
import logging
import amici
import matplotlib as mpl
import numpy as np
import pypesto.optimize as optimize
import pypesto.petab
import pypesto.visualize as visualize
np.random.seed(1)
# increase image resolution
mpl.rcParams["figure.dpi"] = 300
1. Objective Definition
PyPESTO allows the definition of a custom objectives, as well as offers support for objectives defined in the PEtab format.
Custom Objective Definition
You can define an objective via a python function. Also providing an analytical gradient (and potentially also a Hessian) improves the performance of Gradient/Hessian-based optimizers. When accessing parameter uncertainties via profile-likelihoods/sampling, pyPESTO interprets the objective function as the negative-log-likelihood/negative-log-posterior.
[2]:
# define objective function
def f(x: np.array):
return x[0] ** 2 + x[1] ** 2
# define gradient
def grad(x: np.array):
return 2 * x
# define objective
custom_objective = pypesto.Objective(fun=f, grad=grad)
Define lower and upper parameter bounds and create an optimization problem.
[3]:
# define optimization bounds
lb = np.array([-10, -10])
ub = np.array([10, 10])
# create problem
custom_problem = pypesto.Problem(objective=custom_objective, lb=lb, ub=ub)
Now choose an optimizer to perform the optimization. minimize uses multi-start optimization, meaning that the optimization is run n_start times from different initial values, in case the problem contains multiple local optima (which of course is not the case for this toy problem).
[4]:
# choose optimizer
optimizer = optimize.ScipyOptimizer()
# do the optimization
result_custom_problem = optimize.minimize(
problem=custom_problem, optimizer=optimizer, n_starts=10
)
result_custom_problem.optimize_result now stores a list, that contains the results and metadata of the individual optimizer runs (ordered by function value).
[5]:
# E.g., The best model fit was obtained by the following optimization run:
result_custom_problem.optimize_result.list[0]
[5]:
{'id': '1',
'x': array([0., 0.]),
'fval': 0.0,
'grad': array([0., 0.]),
'hess': None,
'res': None,
'sres': None,
'n_fval': 4,
'n_grad': 4,
'n_hess': 0,
'n_res': 0,
'n_sres': 0,
'x0': array([-9.9977125 , -3.95334855]),
'fval0': 115.58322004264032,
'history': <pypesto.history.base.CountHistory at 0x7fae7e4c24d0>,
'exitflag': 0,
'time': 0.0011255741119384766,
'message': 'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL',
'optimizer': "<ScipyOptimizer method=L-BFGS-B options={'disp': False, 'maxfun': 1000}>",
'free_indices': array([0, 1]),
'inner_parameters': None,
'spline_knots': None}
[6]:
# Objective function values of the different optimizer runs:
result_custom_problem.optimize_result.get_for_key("fval")
[6]:
[0.0,
0.0,
0.0,
0.0,
0.0,
0.0,
1.7609224440720174e-36,
3.429973604864721e-36,
6.379533727371235e-36,
8.94847274678846e-36]
Problem Definition via PEtab
Background on PEtab
PyPESTO supports the PEtab standard. PEtab is a data format for specifying parameter estimation problems in systems biology.
A PEtab problem consist of an SBML file, defining the model topology and a set of .tsv files, defining experimental conditions, observables, measurements and parameters (and their optimization bounds, scale, priors…). All files that make up a PEtab problem can be structured in a .yaml file. The pypesto.Objective coming from a PEtab problem corresponds to the negative-log-likelihood/negative-log-posterior distribution of the parameters.
For more details on PEtab, the interested reader is referred to PEtab’s format definition, for examples, the reader is referred to the PEtab benchmark collection. The Model from Böhm et al. JProteomRes 2014 is part of the benchmark collection and will be used as the running example throughout this notebook.
PyPESTO provides an interface to the model simulation tool AMICI for the simulation of Ordinary Differential Equation (ODE) models specified in the SBML format.
Basic Model Import and Optimization
The first step is to import a PEtab problem and create a pypesto.problem object:
[7]:
%%capture
# directory of the PEtab problem
petab_yaml = "./boehm_JProteomeRes2014/boehm_JProteomeRes2014.yaml"
importer = pypesto.petab.PetabImporter.from_yaml(petab_yaml)
problem = importer.create_problem(verbose=False)
Next, we choose an optimizer to perform the multi start optimization.
[8]:
%%time
%%capture
# choose optimizer
optimizer = optimize.ScipyOptimizer()
# do the optimization
result = optimize.minimize(problem=problem, optimizer=optimizer, n_starts=5)
2024-04-16 14:14:15.521 - amici.swig_wrappers - DEBUG - [model1_data1][CVODES:CVode:ERR_FAILURE] AMICI ERROR: in module CVODES in function CVode : At t = 57.5219 and h = 5.38727e-06, the error test failed repeatedly or with |h| = hmin.
2024-04-16 14:14:15.522 - amici.swig_wrappers - ERROR - [model1_data1][FORWARD_FAILURE] AMICI forward simulation failed at t = 57.5219: AMICI failed to integrate the forward problem
CPU times: user 10.7 s, sys: 286 ms, total: 11 s
Wall time: 10.8 s
result.optimize_result contains a list with the ordered optimization results.
[9]:
# E.g., best model fit was obtained by the following optimization run:
result.optimize_result.list[0]
[9]:
{'id': '3',
'x': array([-1.56909438, -5. , -2.20991819, -1.78588311, 2.9545679 ,
4.19767947, 0.693 , 0.58565567, 0.81888913, 0.49860669,
0.107 ]),
'fval': 138.22408495959584,
'grad': array([-0.00233166, 0.05537481, -0.00133531, 0.00174017, -0.00490831,
-0.00381216, nan, -0.00286462, 0.00115818, 0.00010868,
nan]),
'hess': None,
'res': None,
'sres': None,
'n_fval': 190,
'n_grad': 190,
'n_hess': 0,
'n_res': 0,
'n_sres': 0,
'x0': array([ 1.92322616, -3.3016958 , 0.33165285, -4.81711723, 2.89279328,
-2.12224661, 0.693 , -2.34453341, 0.89305537, -0.8582073 ,
0.107 ]),
'fval0': 2175263610.7389483,
'history': <pypesto.history.base.CountHistory at 0x7fae7f7e7850>,
'exitflag': 0,
'time': 5.12551736831665,
'message': 'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH',
'optimizer': "<ScipyOptimizer method=L-BFGS-B options={'disp': False, 'maxfun': 1000}>",
'free_indices': array([0, 1, 2, 3, 4, 5, 7, 8, 9]),
'inner_parameters': None,
'spline_knots': None}
[10]:
# Objective function values of the different optimizer runs:
result.optimize_result.get_for_key("fval")
[10]:
[138.22408495959584,
151.01115875452635,
208.15874132985627,
249.74483886522765,
249.74599744329183]
2. Optimizer Choice
PyPESTO provides a unified interface to a variety of optimizers of different types:
All scipy optimizer (
optimize.ScipyOptimizer(method=<method_name>))function-value or least-squares-based optimizers
gradient or hessian-based optimizers
IpOpt (
optimize.IpoptOptimizer())Interior point method
Dlib (
optimize.DlibOptimizer(options={'maxiter': <max. number of iterations>}))Global optimizer
Gradient-free
FIDES (
optimize.FidesOptimizer())Interior Trust Region optimizer
Particle Swarm (
optimize.PyswarmOptimizer())Particle swarm algorithm
Gradient-free
CMA-ES (
optimize.CmaOptimizer())Covariance Matrix Adaptation Evolution Strategy
Evolutionary Algorithm
[11]:
optimizer_scipy_lbfgsb = optimize.ScipyOptimizer(method="L-BFGS-B")
optimizer_scipy_powell = optimize.ScipyOptimizer(method="Powell")
optimizer_fides = optimize.FidesOptimizer(verbose=logging.ERROR)
optimizer_pyswarm = optimize.PyswarmOptimizer()
The following performs 10 multi-start runs with different optimizers in order to compare their performance. For a faster execution of this notebook, we run only 10 starts. In application, one would use many more optimization starts: around 100-1000 in most cases.
Note: dlib and pyswarm need to be installed for this section to run. Furthermore, the computation time is in the order of minutes, so you might want to skip the execution and jump to the section on large scale models.
[12]:
%%time
%%capture --no-display
n_starts = 10
# Due to run time we already use parallelization.
# This will be introduced in more detail later.
engine = pypesto.engine.MultiProcessEngine()
history_options = pypesto.HistoryOptions(trace_record=True)
# Scipy: L-BFGS-B
result_lbfgsb = optimize.minimize(
problem=problem,
optimizer=optimizer_scipy_lbfgsb,
engine=engine,
history_options=history_options,
n_starts=n_starts,
)
# Scipy: Powell
result_powell = optimize.minimize(
problem=problem,
optimizer=optimizer_scipy_powell,
engine=engine,
history_options=history_options,
n_starts=n_starts,
)
# Fides
result_fides = optimize.minimize(
problem=problem,
optimizer=optimizer_fides,
engine=engine,
history_options=history_options,
n_starts=n_starts,
)
# PySwarm
result_pyswarm = optimize.minimize(
problem=problem,
optimizer=optimizer_pyswarm,
engine=engine,
history_options=history_options,
n_starts=n_starts,
)
Engine will use up to 2 processes (= CPU count).
2024-04-16 14:14:33.761 - amici.swig_wrappers - DEBUG - [model1_data1][CVODES:CVode:ERR_FAILURE] AMICI ERROR: in module CVODES in function CVode : At t = 14.1895 and h = 4.87286e-06, the error test failed repeatedly or with |h| = hmin.
2024-04-16 14:14:33.767 - amici.swig_wrappers - ERROR - [model1_data1][FORWARD_FAILURE] AMICI forward simulation failed at t = 14.1895: AMICI failed to integrate the forward problem
2024-04-16 14:14:33.863 - amici.swig_wrappers - DEBUG - [model1_data1][CVODES:CVode:ERR_FAILURE] AMICI ERROR: in module CVODES in function CVode : At t = 14.1895 and h = 4.87286e-06, the error test failed repeatedly or with |h| = hmin.
2024-04-16 14:14:33.867 - amici.swig_wrappers - ERROR - [model1_data1][FORWARD_FAILURE] AMICI forward simulation failed at t = 14.1895: AMICI failed to integrate the forward problem
2024-04-16 14:14:58.370 - amici.swig_wrappers - DEBUG - [model1_data1][CVODES:CVode:ERR_FAILURE] AMICI ERROR: in module CVODES in function CVode : At t = 91.9897 and h = 6.68112e-06, the error test failed repeatedly or with |h| = hmin.
2024-04-16 14:14:58.376 - amici.swig_wrappers - ERROR - [model1_data1][FORWARD_FAILURE] AMICI forward simulation failed at t = 91.9897: AMICI failed to integrate the forward problem
2024-04-16 14:15:01.858 - amici.swig_wrappers - DEBUG - [model1_data1][CVODES:CVode:ERR_FAILURE] AMICI ERROR: in module CVODES in function CVode : At t = 145.61 and h = 2.2797e-05, the error test failed repeatedly or with |h| = hmin.
2024-04-16 14:15:01.862 - amici.swig_wrappers - ERROR - [model1_data1][FORWARD_FAILURE] AMICI forward simulation failed at t = 145.61: AMICI failed to integrate the forward problem
2024-04-16 14:15:03.783 - amici.swig_wrappers - DEBUG - [model1_data1][CVODES:CVode:ERR_FAILURE] AMICI ERROR: in module CVODES in function CVode : At t = 145.018 and h = 6.81561e-06, the error test failed repeatedly or with |h| = hmin.
2024-04-16 14:15:03.787 - amici.swig_wrappers - ERROR - [model1_data1][FORWARD_FAILURE] AMICI forward simulation failed at t = 145.018: AMICI failed to integrate the forward problem
2024-04-16 14:15:05.396 - amici.swig_wrappers - DEBUG - [model1_data1][CVODES:CVode:ERR_FAILURE] AMICI ERROR: in module CVODES in function CVode : At t = 145.61 and h = 3.08081e-05, the error test failed repeatedly or with |h| = hmin.
2024-04-16 14:15:05.398 - amici.swig_wrappers - ERROR - [model1_data1][FORWARD_FAILURE] AMICI forward simulation failed at t = 145.61: AMICI failed to integrate the forward problem
CPU times: user 684 ms, sys: 117 ms, total: 801 ms
Wall time: 3min 21s
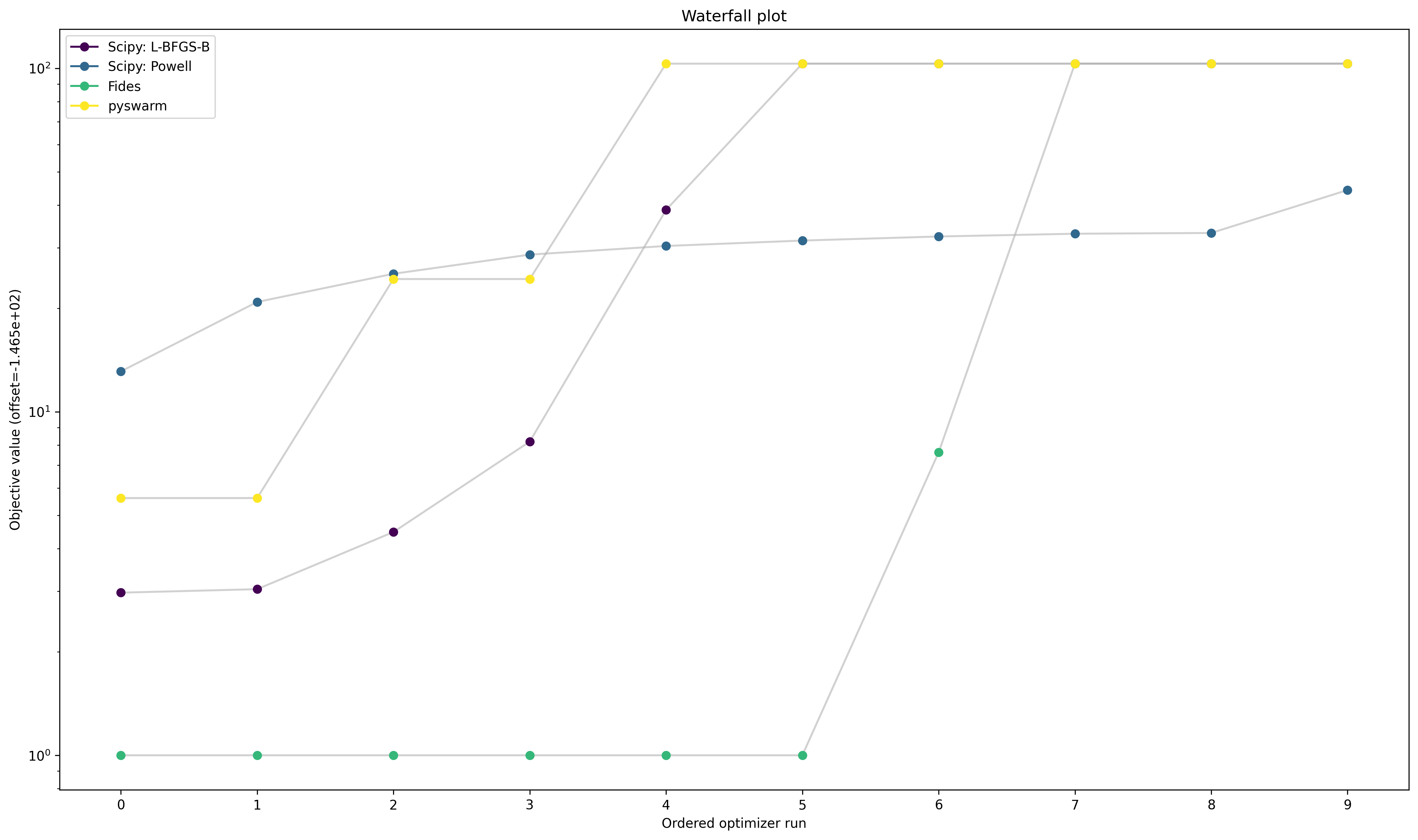
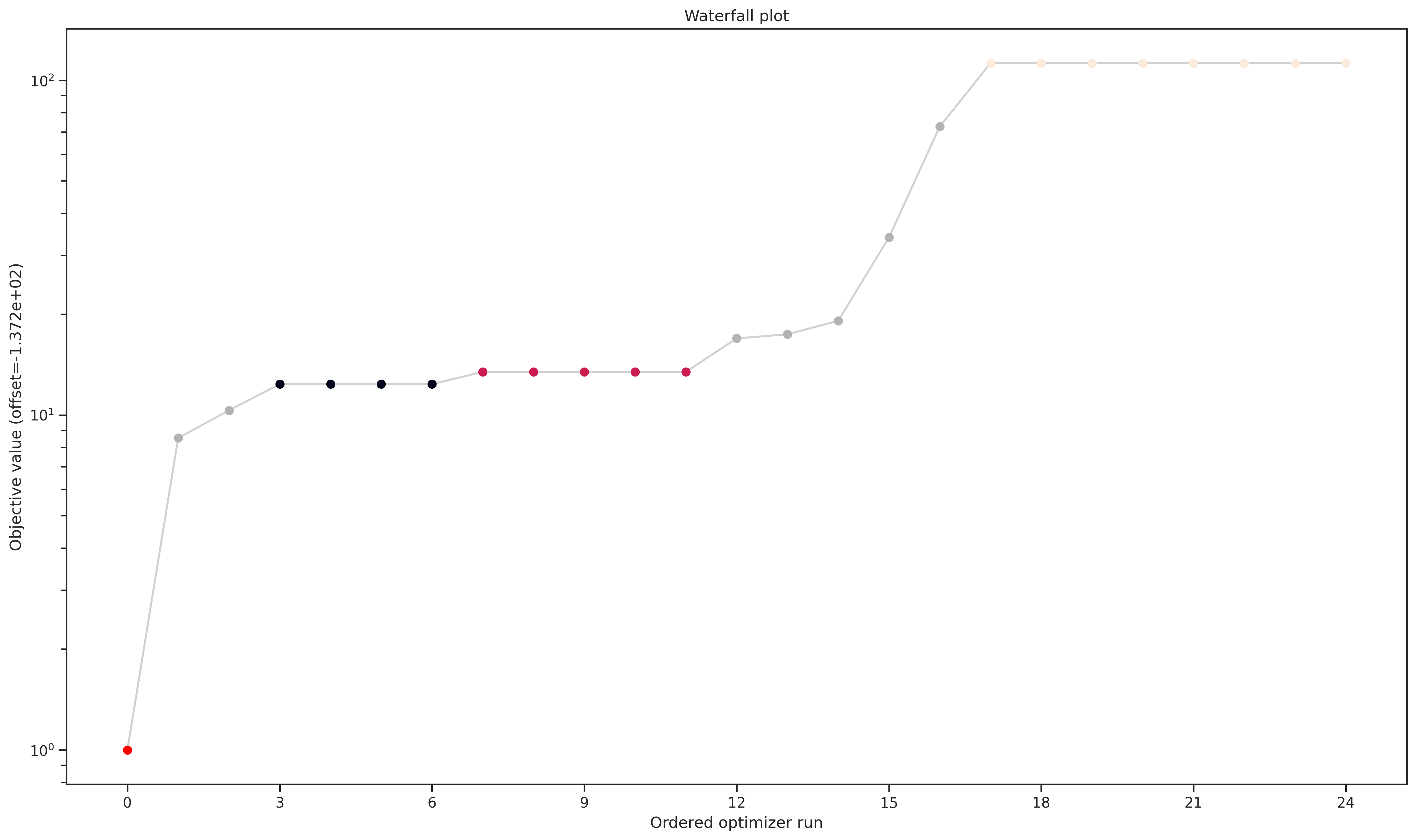
Optimizer Convergence
A common visualization of optimizer convergence are waterfall plots. Waterfall plots show the (ordered) results of the individual optimization runs. As we see below, Dlib and pyswarm, which are not gradient-based, are not able to find the global optimum.
Furthermore, we hope to obtain clearly visible plateaus, as they indicate optimizer convergence to local minima.
[13]:
optimizer_results = [
result_lbfgsb,
result_powell,
result_fides,
result_pyswarm,
]
optimizer_names = ["Scipy: L-BFGS-B", "Scipy: Powell", "Fides", "pyswarm"]
pypesto.visualize.waterfall(optimizer_results, legends=optimizer_names);
Optimizer run time
Optimizer run time vastly differs among the different optimizers, as can be seen below:
[14]:
print("Average Run time per start:")
print("-------------------")
for optimizer_name, optimizer_result in zip(
optimizer_names, optimizer_results
):
t = np.sum(optimizer_result.optimize_result.get_for_key("time")) / n_starts
print(f"{optimizer_name}: {t:f} s")
Average Run time per start:
-------------------
Scipy: L-BFGS-B: 3.233474 s
Scipy: Powell: 3.383111 s
Fides: 1.645531 s
pyswarm: 31.222015 s
3. Fitting of large scale models
When fitting large scale models (i.e. with >100 parameters and accordingly also more data), two important issues are efficient gradient computation and parallelization.
Efficient gradient computation
As seen in the example above and as can be confirmed from own experience: If fast and reliable gradients can be provided, gradient-based optimizers are favourable with respect to optimizer convergence and run time.
It has been shown that adjoint sensitivity analysis is a fast and reliable method to compute gradients for large scale models, since their run time is (asymptotically) independent of the number of parameters (Fröhlich et al. PlosCB 2017).
(Figure from Fröhlich et al. PlosCB 2017) Adjoint sensitivities are implemented in AMICI.
[15]:
# Set gradient computation method to adjoint
problem.objective.amici_solver.setSensitivityMethod(
amici.SensitivityMethod.adjoint
)
Parallelization
Multi-start optimization can easily be parallelized by using engines.
[16]:
%%time
%%capture
# Parallelize
engine = pypesto.engine.MultiProcessEngine()
# Optimize
result = optimize.minimize(
problem=problem,
optimizer=optimizer_scipy_lbfgsb,
engine=engine,
n_starts=25,
)
Engine will use up to 2 processes (= CPU count).
CPU times: user 159 ms, sys: 42.8 ms, total: 202 ms
Wall time: 1min 34s
4. Uncertainty quantification
PyPESTO focuses on two approaches to assess parameter uncertainties:
Profile likelihoods
Sampling
Profile Likelihoods
Profile likelihoods compute confidence intervals via a likelihood ratio test. Profile likelihoods perform a maximum-projection of the likelihood function on the parameter of interest. The likelihood ratio test then gives a cut-off criterion via the \(\chi^2_1\) distribution.
In pyPESTO, the maximum projection is solved as a maximization problem and can be obtained via
[17]:
%%time
%%capture
import pypesto.profile as profile
result = profile.parameter_profile(
problem=problem,
result=result,
optimizer=optimizer_scipy_lbfgsb,
profile_index=[0, 1, 2],
)
CPU times: user 33 s, sys: 687 ms, total: 33.7 s
Wall time: 33.3 s
The maximum projections can now be inspected via:
[18]:
# adapt x_labels..
x_labels = [f"Log10({name})" for name in problem.x_names]
visualize.profiles(result, x_labels=x_labels, show_bounds=True);
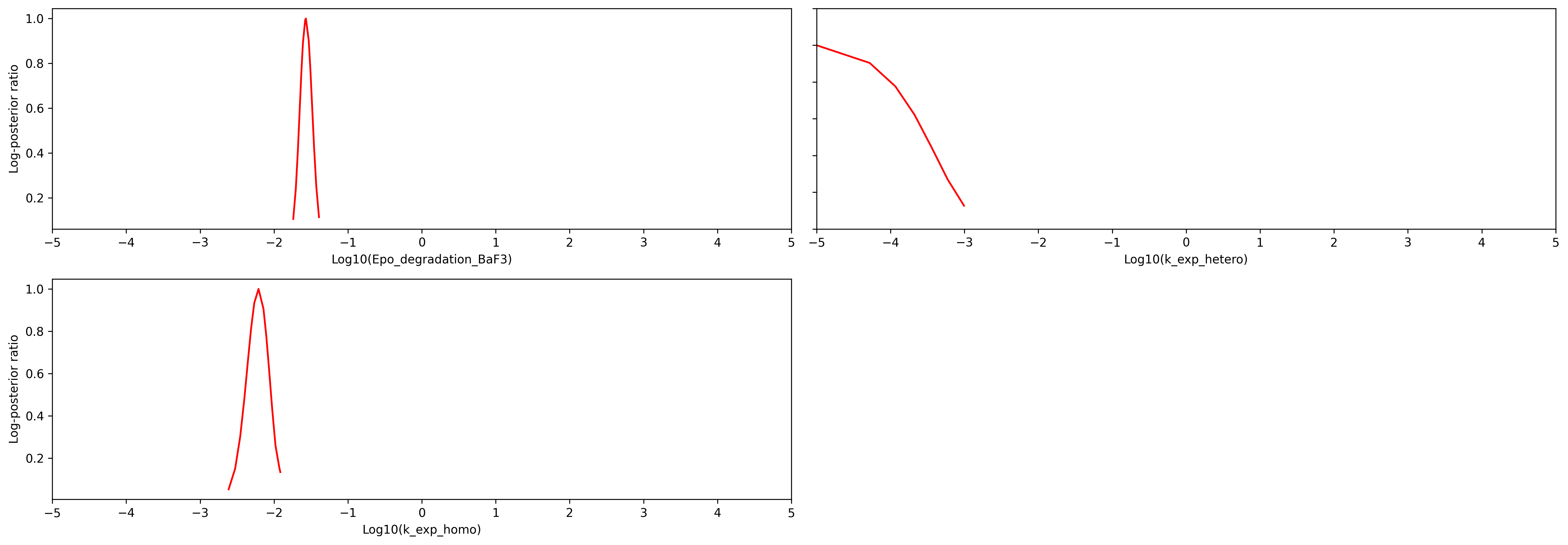
The plot shows that seven parameters are identifiable, since the likelihood is tightly centered around the optimal parameter. Two parameters (k_exp_hetero and k_imp_homo) cannot be constrained by the data.
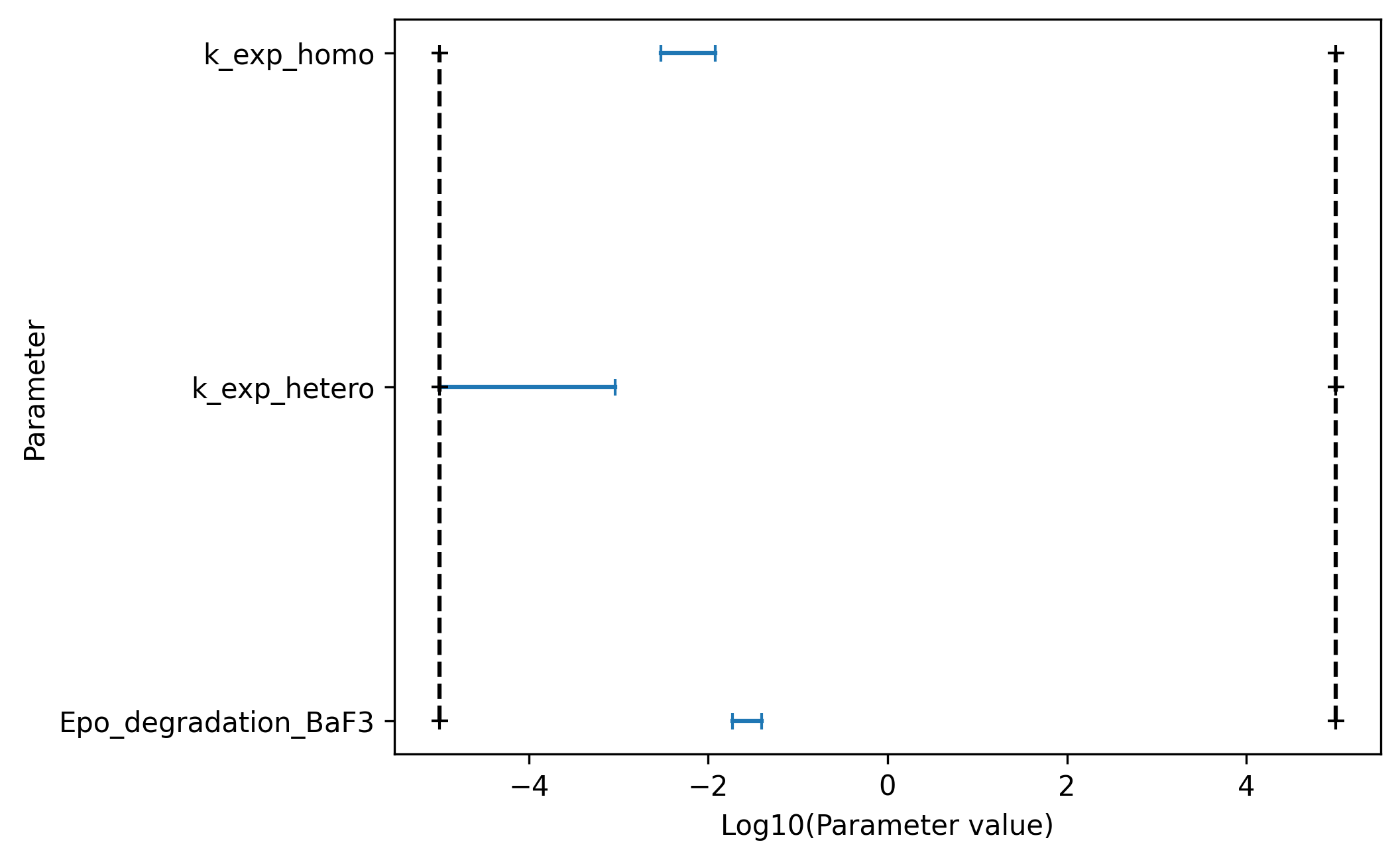
Furthermore pyPESTO allows to visualize confidence intervals directly via
[19]:
ax = pypesto.visualize.profile_cis(
result, confidence_level=0.95, show_bounds=True
)
ax.set_xlabel("Log10(Parameter value)");
Sampling
In pyPESTO, sampling from the posterior distribution can be performed as
[20]:
import pypesto.sample as sample
n_samples = 1000
sampler = sample.AdaptiveMetropolisSampler()
result = sample.sample(
problem, n_samples=n_samples, sampler=sampler, result=result
)
Elapsed time: 2.8500575659999967
Sampling results are stored in result.sample_result and can be accessed e.g., via
[21]:
result.sample_result["trace_x"]
[21]:
array([[[-1.56904137, -5. , -2.20989351, ..., 0.58567235,
0.81883295, 0.49858573],
[-1.56904137, -5. , -2.20989351, ..., 0.58567235,
0.81883295, 0.49858573],
[-1.56904137, -5. , -2.20989351, ..., 0.58567235,
0.81883295, 0.49858573],
...,
[-1.60750199, -4.8263492 , -2.19080131, ..., 0.5500663 ,
0.76865892, 0.52492182],
[-1.61746266, -4.80609266, -2.17959698, ..., 0.55721972,
0.77760402, 0.53645899],
[-1.61746266, -4.80609266, -2.17959698, ..., 0.55721972,
0.77760402, 0.53645899]]])
Sampling Diagnostics
Geweke’s test assesses convergence of a sampling run and computes the burn-in of a sampling result. The effective sample size indicates the strength of the correlation between different samples.
[22]:
sample.geweke_test(result=result)
result.sample_result["burn_in"]
Geweke burn-in index: 0
[22]:
0
[23]:
sample.effective_sample_size(result=result)
result.sample_result["effective_sample_size"]
Estimated chain autocorrelation: 94.74853956063886
Estimated effective sample size: 10.454467552124417
[23]:
10.454467552124417
Visualization of Sampling Results
[24]:
# scatter plots
visualize.sampling_scatter(result)
# marginals
visualize.sampling_1d_marginals(result);
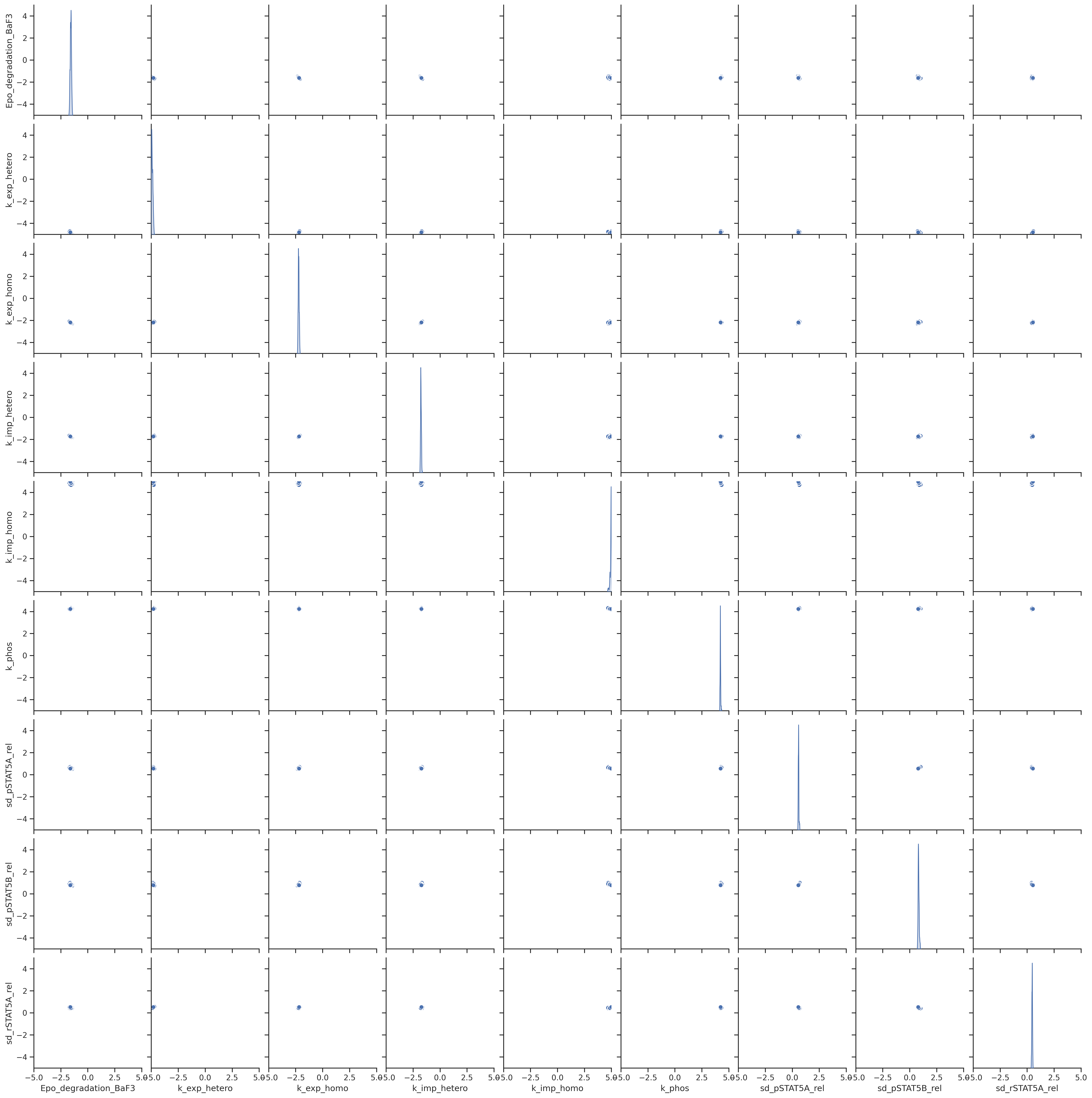
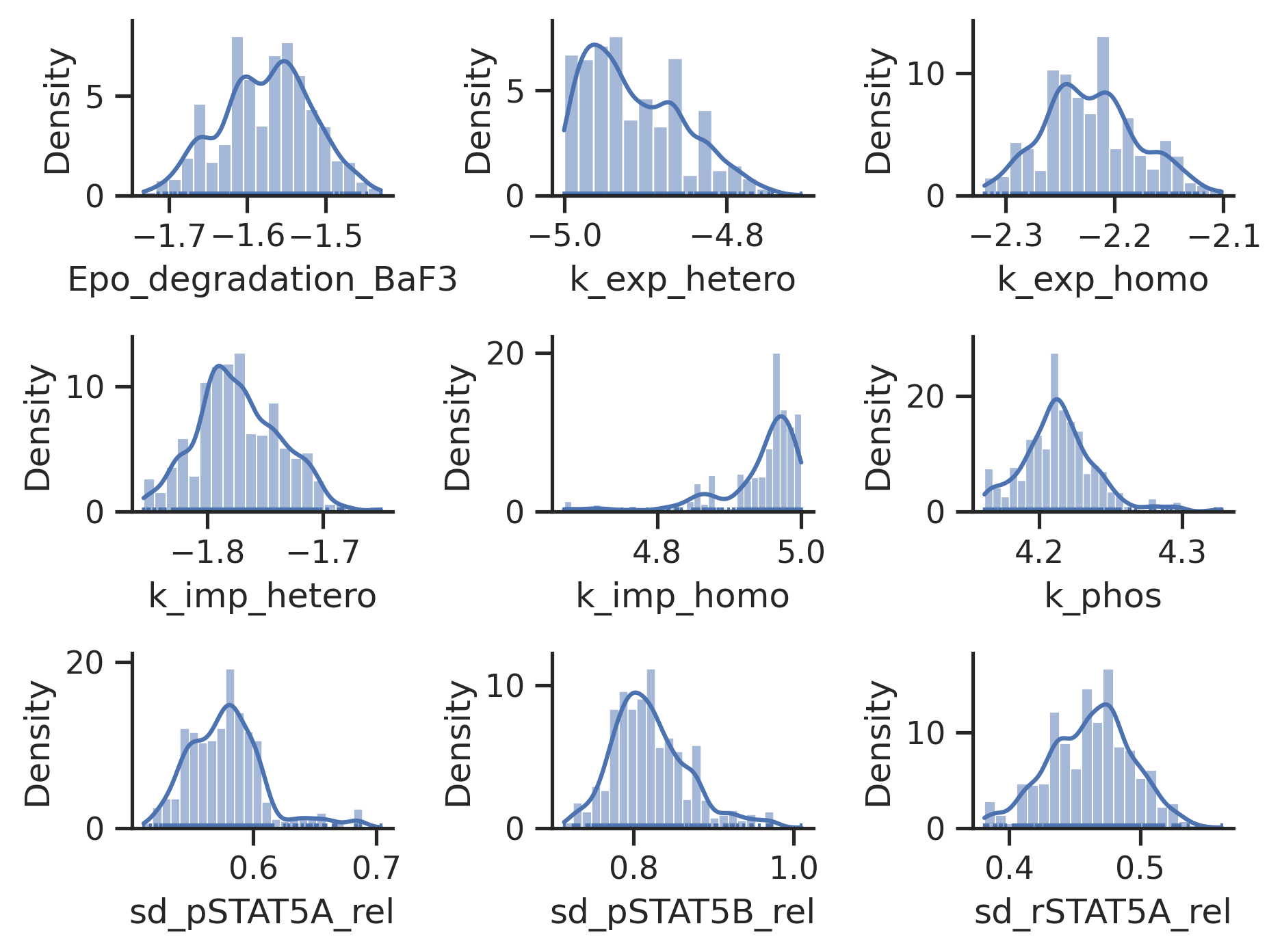
Sampler Choice:
Similarly to parameter optimization, pyPESTO provides a unified interface to several sampler/sampling toolboxes, as well as own implementations of sampler:
Adaptive Metropolis:
sample.AdaptiveMetropolisSampler()Adaptive parallel tempering:
sample.ParallelTemperingSampler()Interface to
pymc3viasample.Pymc3Sampler()
5. Storage
You can store and load the results of an analysis via the pypesto.store module to a .hdf5 file.
Store result
[25]:
import tempfile
import pypesto.store as store
# create a temporary file, for demonstration purpose
f_tmp = tempfile.NamedTemporaryFile(suffix=".hdf5", delete=False)
result_file_name = f_tmp.name
# store the result
store.write_result(result, result_file_name)
f_tmp.close()
Load result file
You can re-load a result, e.g. for visualizations:
[26]:
# read result
result_loaded = store.read_result(result_file_name)
# e.g. do some visualisation
visualize.waterfall(result_loaded);
/home/docs/checkouts/readthedocs.org/user_builds/pypesto/envs/latest/lib/python3.11/site-packages/pypesto/store/read_from_hdf5.py:304: UserWarning: You are loading a problem. This problem is not to be used without a separately created objective.
result.problem = pypesto_problem_reader.read()
Software Development Standards:
PyPESTO is developed with the following standards:
Open source, code on GitHub.
Pip installable via:
pip install pypesto.Documentation as RTD and example jupyter notebooks are available.
Has continuous integration & extensive automated testing.
Code reviews before merging into the develop/main branch.
Currently, 5–10 people are using, extending and (most importantly) maintaining pyPESTO in their “daily business”.
Further topics
Further features are available, among them:
Model Selection
Hierarchical Optimization of scaling/noise parameters
Categorical Data