Rosenbrock banana¶
Here, we perform optimization for the Rosenbrock banana function, which does not require an AMICI model. In particular, we try several ways of specifying derivative information.
[1]:
# install if not done yet
# %pip install pypesto --quiet
[2]:
import matplotlib.pyplot as plt
import numpy as np
import scipy as sp
from mpl_toolkits.mplot3d import Axes3D
import pypesto
import pypesto.visualize as visualize
%matplotlib inline
Define the objective and problem¶
[3]:
# first type of objective
objective1 = pypesto.Objective(
fun=sp.optimize.rosen,
grad=sp.optimize.rosen_der,
hess=sp.optimize.rosen_hess,
)
# second type of objective
def rosen2(x):
return (
sp.optimize.rosen(x),
sp.optimize.rosen_der(x),
sp.optimize.rosen_hess(x),
)
objective2 = pypesto.Objective(fun=rosen2, grad=True, hess=True)
dim_full = 10
lb = -5 * np.ones((dim_full, 1))
ub = 5 * np.ones((dim_full, 1))
problem1 = pypesto.Problem(objective=objective1, lb=lb, ub=ub)
problem2 = pypesto.Problem(objective=objective2, lb=lb, ub=ub)
Illustration¶
[4]:
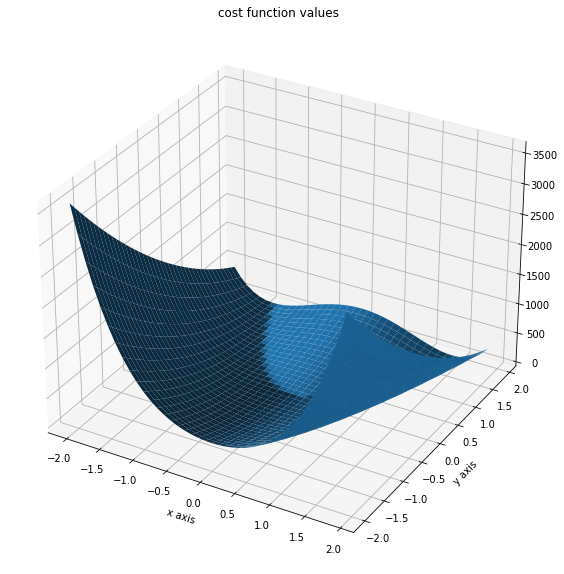
x = np.arange(-2, 2, 0.1)
y = np.arange(-2, 2, 0.1)
x, y = np.meshgrid(x, y)
z = np.zeros_like(x)
for j in range(0, x.shape[0]):
for k in range(0, x.shape[1]):
z[j, k] = objective1([x[j, k], y[j, k]], (0,))
[5]:
fig = plt.figure()
fig.set_size_inches(*(14, 10))
ax = plt.axes(projection="3d")
ax.plot_surface(X=x, Y=y, Z=z)
plt.xlabel("x axis")
plt.ylabel("y axis")
ax.set_title("cost function values")
[5]:
Text(0.5, 0.92, 'cost function values')
Run optimization¶
[6]:
import pypesto.optimize as optimize
[7]:
# create different optimizers
optimizer_bfgs = optimize.ScipyOptimizer(method="l-bfgs-b")
optimizer_tnc = optimize.ScipyOptimizer(method="TNC")
optimizer_dogleg = optimize.ScipyOptimizer(method="dogleg")
# set number of starts
n_starts = 20
# save optimizer trace
history_options = pypesto.HistoryOptions(trace_record=True)
# Run optimizaitons for different optimzers
result1_bfgs = optimize.minimize(
problem=problem1,
optimizer=optimizer_bfgs,
n_starts=n_starts,
history_options=history_options,
filename=None,
)
result1_tnc = optimize.minimize(
problem=problem1,
optimizer=optimizer_tnc,
n_starts=n_starts,
history_options=history_options,
filename=None,
)
result1_dogleg = optimize.minimize(
problem=problem1,
optimizer=optimizer_dogleg,
n_starts=n_starts,
history_options=history_options,
filename=None,
)
# Optimize second type of objective
result2 = optimize.minimize(
problem=problem2, optimizer=optimizer_tnc, n_starts=n_starts, filename=None
)
100%|█████████████████████████████████████████████████████████████████| 20/20 [00:00<00:00, 23.57it/s]
100%|█████████████████████████████████████████████████████████████████| 20/20 [00:01<00:00, 13.47it/s]
100%|████████████████████████████████████████████████████████████████| 20/20 [00:00<00:00, 259.22it/s]
100%|█████████████████████████████████████████████████████████████████| 20/20 [00:01<00:00, 10.49it/s]
Visualize and compare optimization results¶
[8]:
# plot separated waterfalls
visualize.waterfall(result1_bfgs, size=(15, 6))
visualize.waterfall(result1_tnc, size=(15, 6))
visualize.waterfall(result1_dogleg, size=(15, 6));
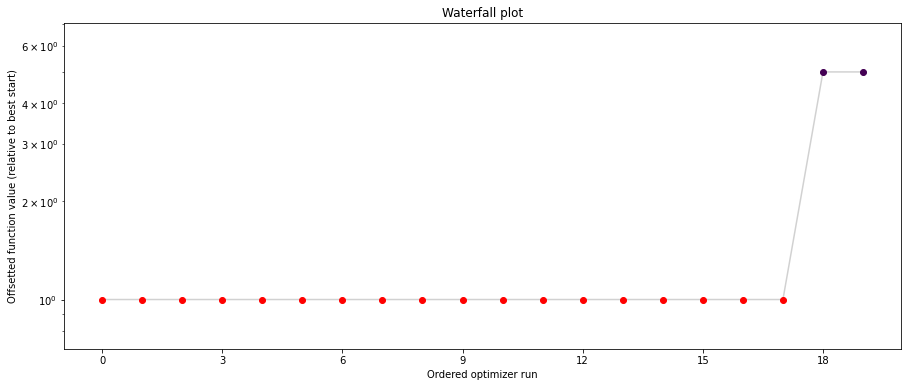
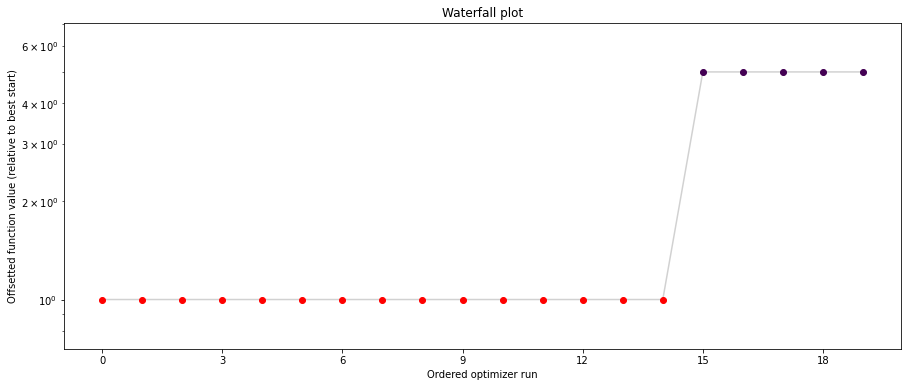
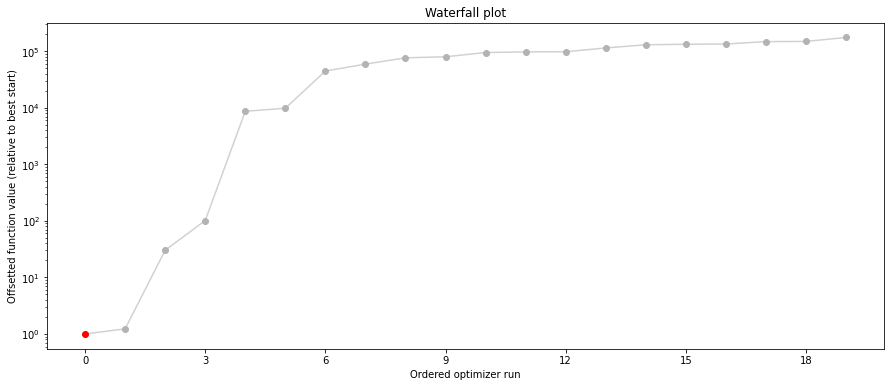
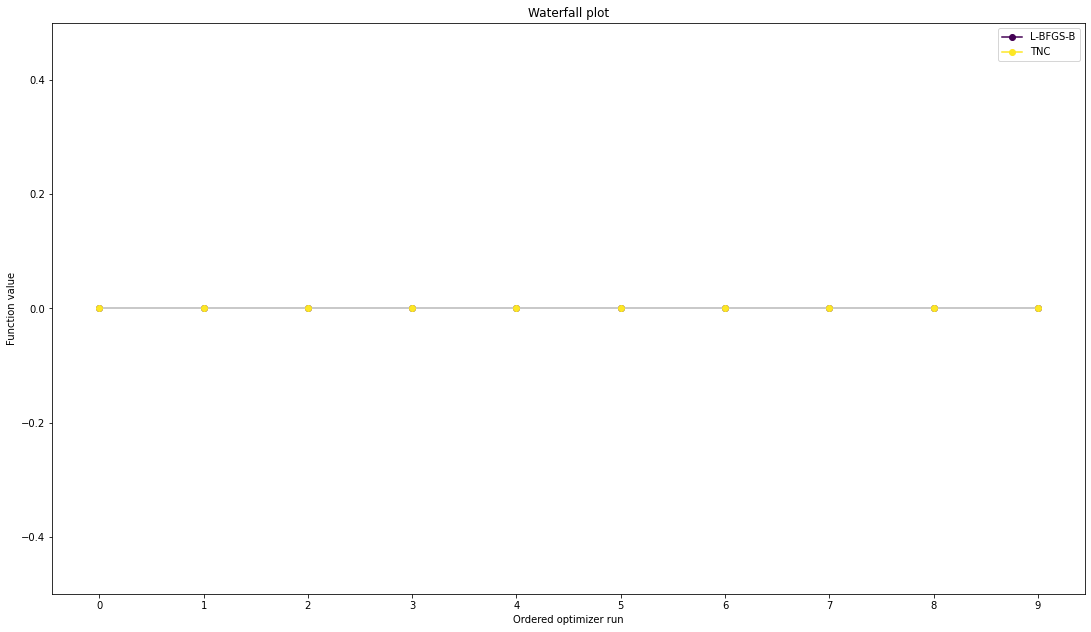
We can now have a closer look, which method perfomred better: Let’s first compare bfgs and TNC, since both methods gave good results. How does the fine convergence look like?
[9]:
# plot one list of waterfalls
visualize.waterfall(
[result1_bfgs, result1_tnc],
legends=["L-BFGS-B", "TNC"],
start_indices=10,
scale_y="lin",
);
[10]:
# retrieve second optimum
all_x = result1_bfgs.optimize_result.x
all_fval = result1_bfgs.optimize_result.fval
x = all_x[19]
fval = all_fval[19]
print("Second optimum at: " + str(fval))
# create a reference point from it
ref = {
"x": x,
"fval": fval,
"color": [0.2, 0.4, 1.0, 1.0],
"legend": "second optimum",
}
ref = visualize.create_references(ref)
# new waterfall plot with reference point for second optimum
visualize.waterfall(
result1_dogleg,
size=(15, 6),
scale_y="lin",
y_limits=[-1, 101],
reference=ref,
colors=[0.0, 0.0, 0.0, 1.0],
);
Second optimum at: 3.986579112988829
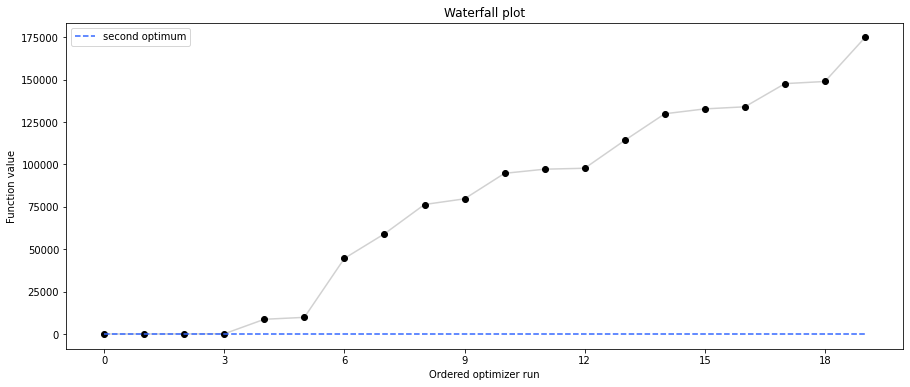
Visualize parameters¶
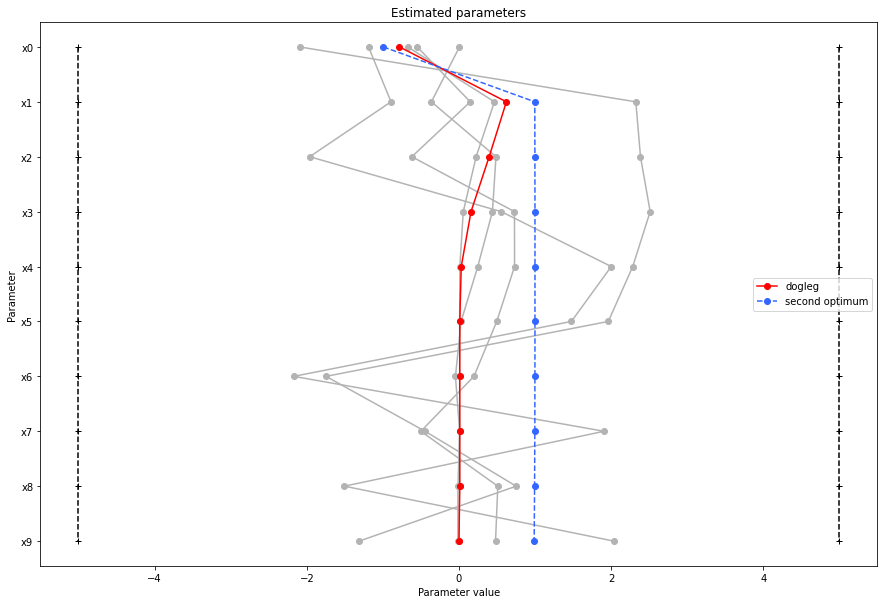
There seems to be a second local optimum. We want to see whether it was also found by the dogleg method
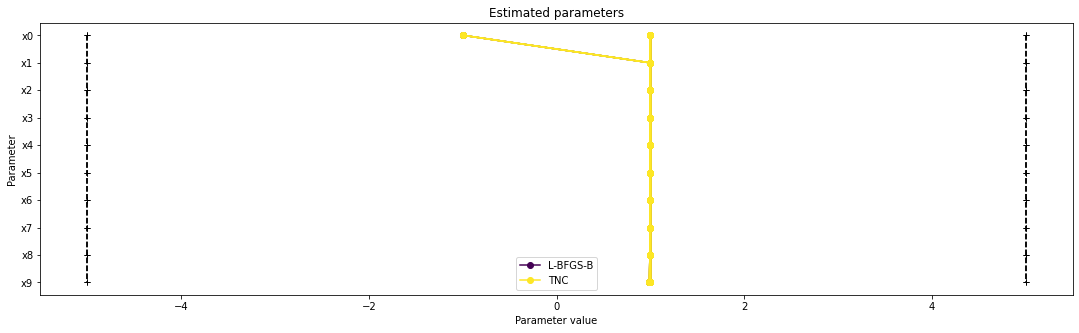
[11]:
visualize.parameters(
[result1_bfgs, result1_tnc],
legends=["L-BFGS-B", "TNC"],
balance_alpha=False,
)
visualize.parameters(
result1_dogleg,
legends="dogleg",
reference=ref,
size=(15, 10),
start_indices=[0, 1, 2, 3, 4, 5],
balance_alpha=False,
);
If the result needs to be examined in more detail, it can easily be exported as a pandas.DataFrame:
[12]:
df = result1_tnc.optimize_result.as_dataframe(
["fval", "n_fval", "n_grad", "n_hess", "n_res", "n_sres", "time"],
)
df.head()
[12]:
| fval | n_fval | n_grad | n_hess | n_res | n_sres | time | |
|---|---|---|---|---|---|---|---|
| 0 | 1.700177e-12 | 234 | 234 | 0 | 0 | 0 | 0.095119 |
| 1 | 5.863075e-12 | 254 | 254 | 0 | 0 | 0 | 0.102610 |
| 2 | 6.024982e-12 | 201 | 201 | 0 | 0 | 0 | 0.074668 |
| 3 | 1.022637e-11 | 231 | 231 | 0 | 0 | 0 | 0.086636 |
| 4 | 1.491191e-11 | 170 | 170 | 0 | 0 | 0 | 0.072328 |
Optimizer history¶
Let’s compare optimzer progress over time.
[13]:
# plot one list of waterfalls
visualize.optimizer_history(
[result1_bfgs, result1_tnc],
legends=["L-BFGS-B", "TNC"],
reference=ref,
)
# plot one list of waterfalls
visualize.optimizer_history(
result1_dogleg,
reference=ref,
);
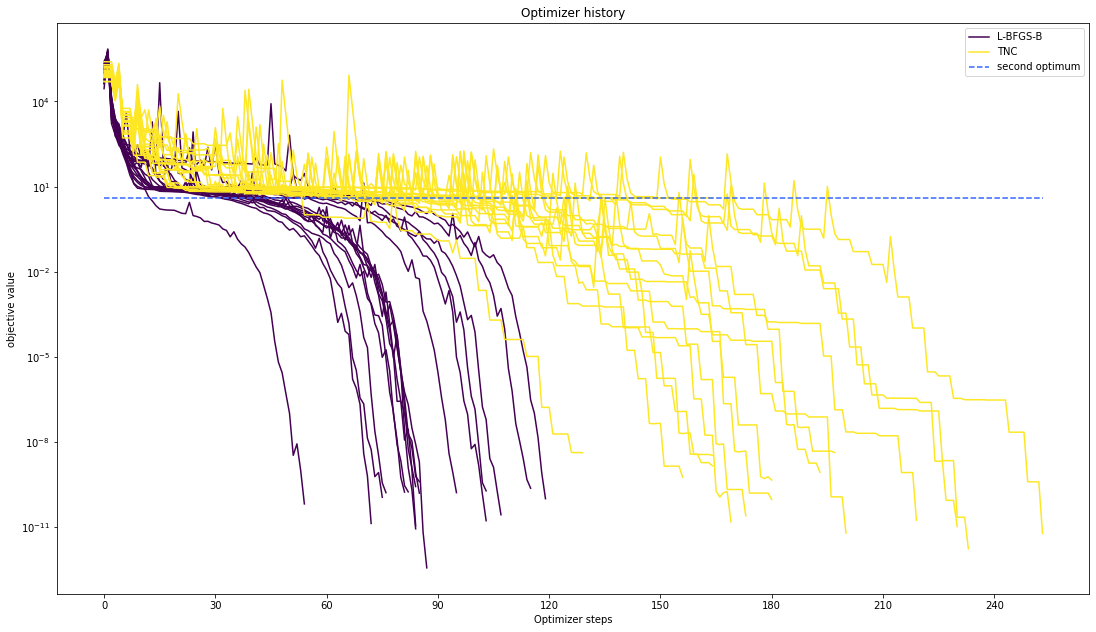
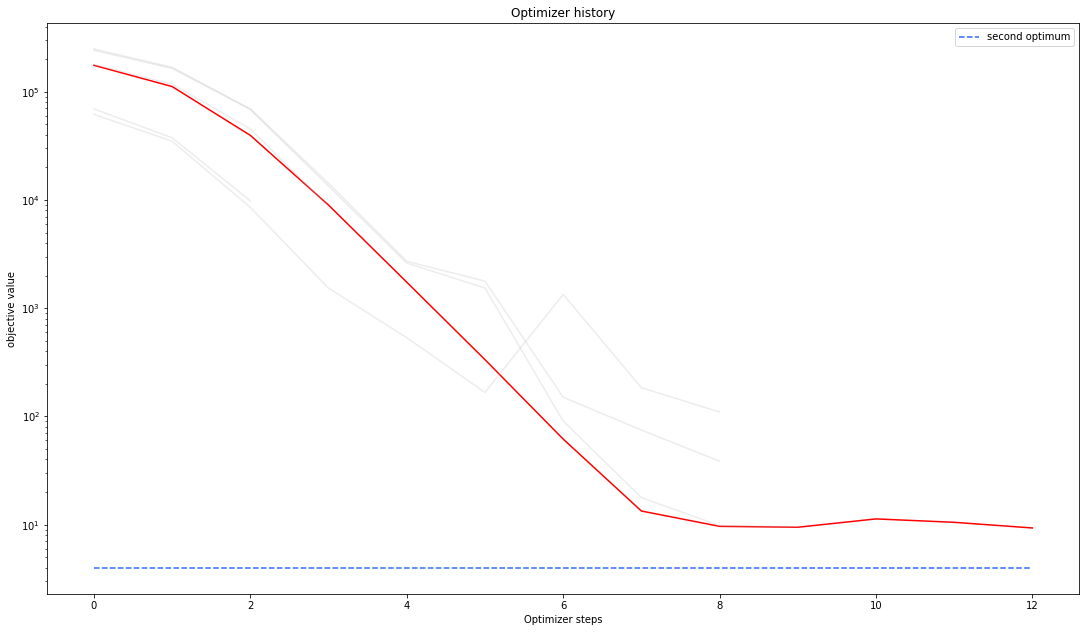
We can also visualize this usign other scalings or offsets…
[14]:
# plot one list of waterfalls
visualize.optimizer_history(
[result1_bfgs, result1_tnc],
legends=["L-BFGS-B", "TNC"],
reference=ref,
offset_y=0.0,
)
# plot one list of waterfalls
visualize.optimizer_history(
[result1_bfgs, result1_tnc],
legends=["L-BFGS-B", "TNC"],
reference=ref,
scale_y="lin",
y_limits=[-1.0, 11.0],
);
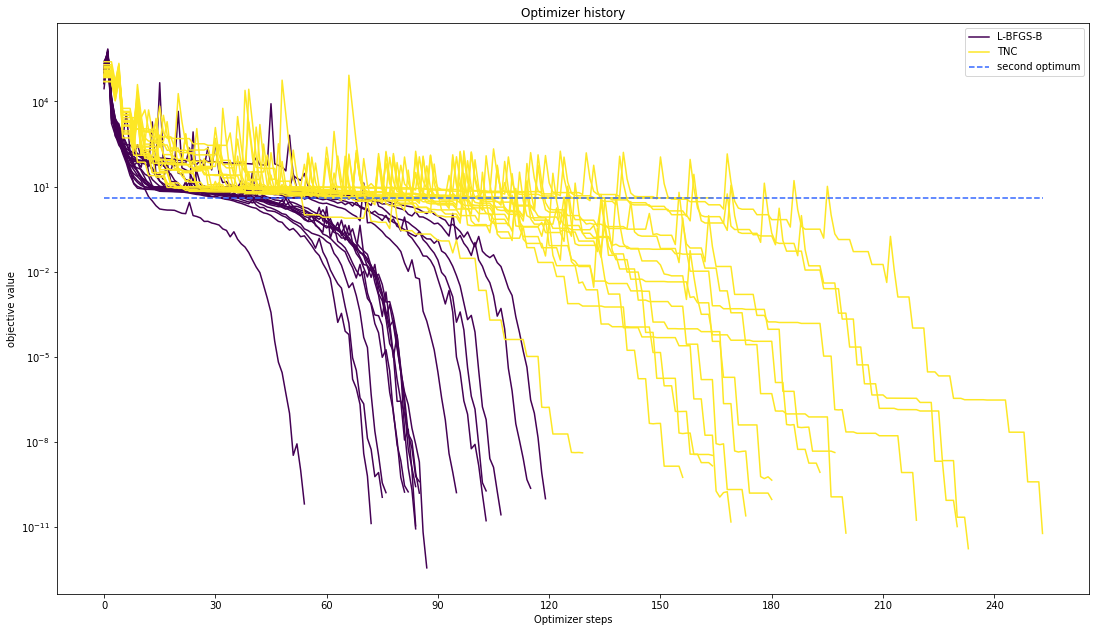
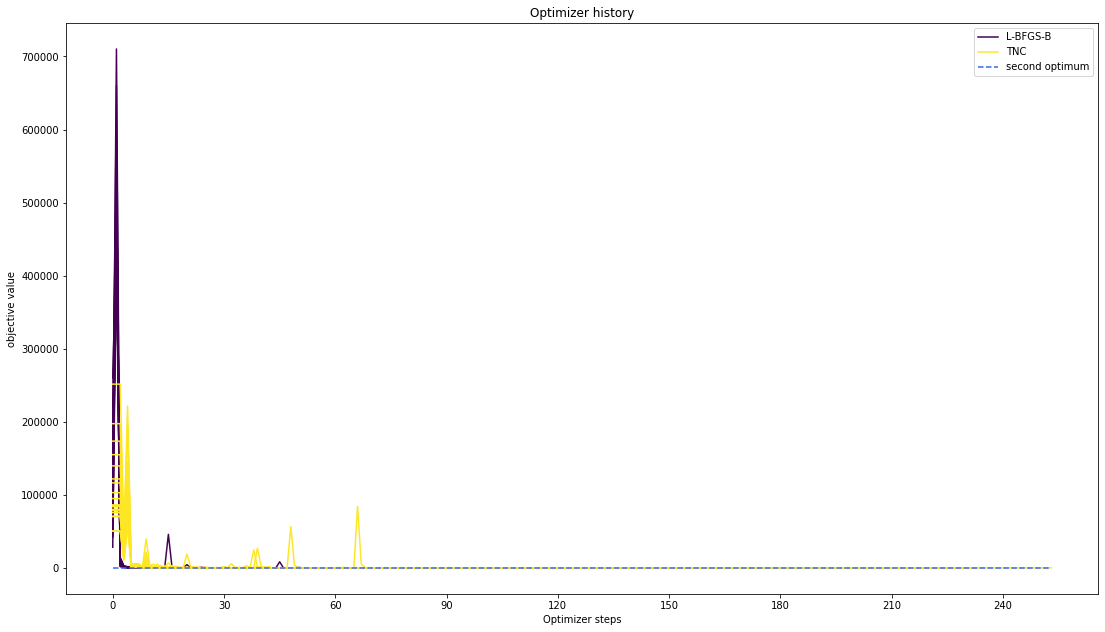
Compute profiles¶
The profiling routine needs a problem, a results object and an optimizer.
Moreover it accepts an index of integer (profile_index), whether or not a profile should be computed.
Finally, an integer (result_index) can be passed, in order to specify the local optimum, from which profiling should be started.
[15]:
import pypesto.profile as profile
[16]:
# compute profiles
profile_options = profile.ProfileOptions(
min_step_size=0.0005,
delta_ratio_max=0.05,
default_step_size=0.005,
ratio_min=0.01,
)
result1_bfgs = profile.parameter_profile(
problem=problem1,
result=result1_bfgs,
optimizer=optimizer_bfgs,
profile_index=np.array([1, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0]),
result_index=0,
profile_options=profile_options,
filename=None,
)
# compute profiles from second optimum
result1_bfgs = profile.parameter_profile(
problem=problem1,
result=result1_bfgs,
optimizer=optimizer_bfgs,
profile_index=np.array([1, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0]),
result_index=19,
profile_options=profile_options,
filename=None,
)
100%|█████████████████████████████████████████████████████████████████| 11/11 [00:00<00:00, 27.63it/s]
100%|█████████████████████████████████████████████████████████████████| 11/11 [00:00<00:00, 86.06it/s]
Visualize and analyze results¶
pypesto offers easy-to-use visualization routines:
[17]:
# specify the parameters, for which profiles should be computed
ax = visualize.profiles(
result1_bfgs,
profile_indices=[0, 1, 2, 5, 7],
reference=ref,
profile_list_ids=[0, 1],
);
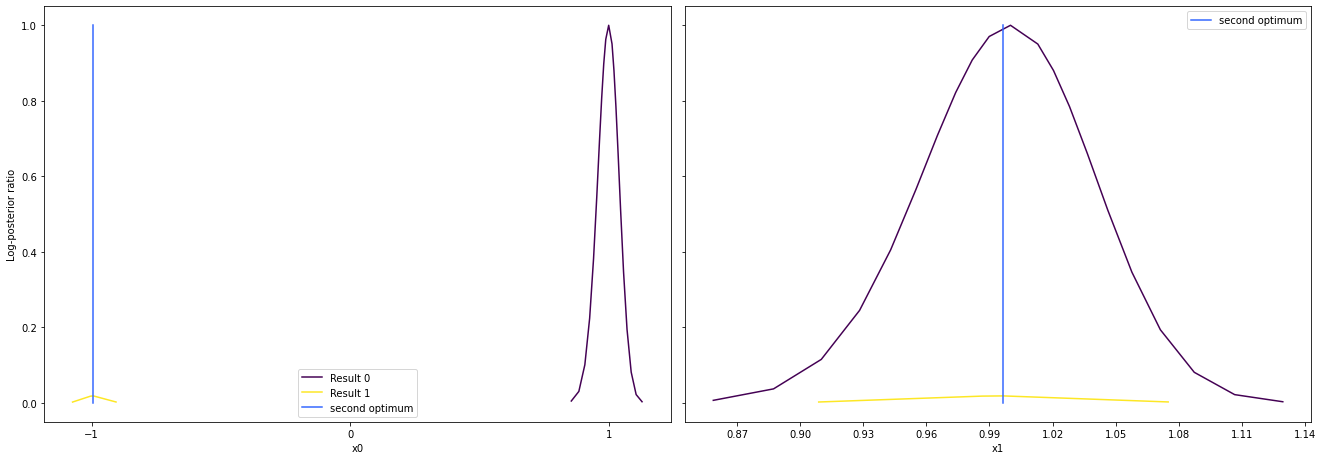
Approximate profiles¶
When computing the profiles is computationally too demanding, it is possible to employ to at least consider a normal approximation with covariance matrix given by the Hessian or FIM at the optimal parameters.
[18]:
result1_tnc = profile.approximate_parameter_profile(
problem=problem1,
result=result1_bfgs,
profile_index=np.array([1, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0]),
result_index=0,
n_steps=1000,
)
Computing Hessian/FIM as not available in result.
These approximate profiles require at most one additional function evaluation, can however yield substantial approximation errors:
[19]:
axes = visualize.profiles(
result1_bfgs,
profile_indices=[0, 1, 2, 5, 7],
profile_list_ids=[0, 2],
ratio_min=0.01,
colors=[(1, 0, 0, 1), (0, 0, 1, 1)],
legends=[
"Optimization-based profile",
"Local profile approximation",
],
);
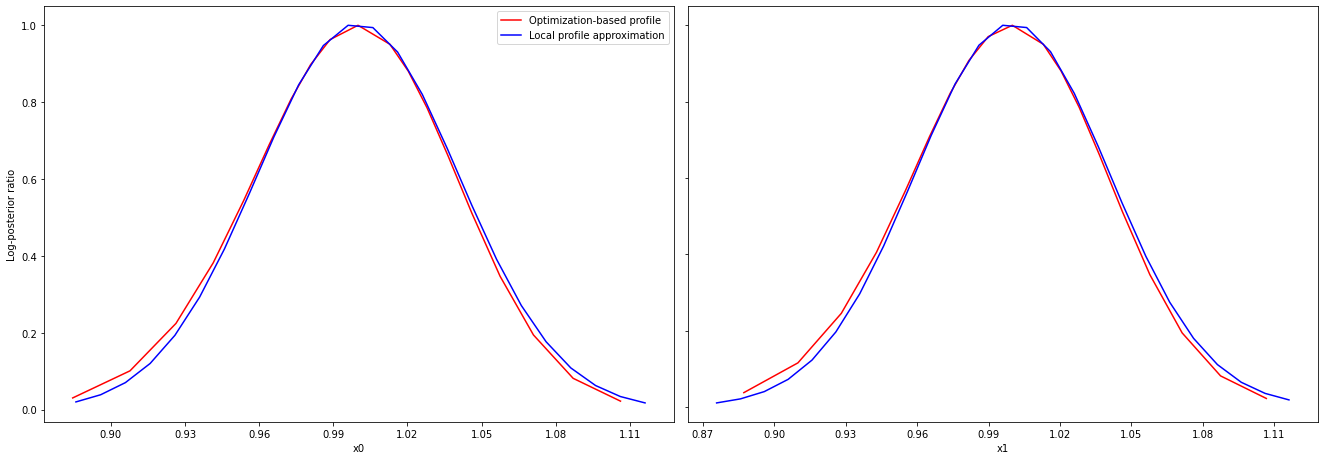
We can also plot approximate confidence intervals based on profiles:
[20]:
visualize.profile_cis(
result1_bfgs,
confidence_level=0.95,
profile_list=2,
)
[20]:
<AxesSubplot:xlabel='Parameter value', ylabel='Parameter'>
