Rosenbrock banana¶
Here, we perform optimization for the Rosenbrock banana function, which does not require an AMICI model. In particular, we try several ways of specifying derivative information.
[1]:
import pypesto
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
%matplotlib inline
Define the objective and problem¶
[2]:
# first type of objective
objective1 = pypesto.Objective(fun=sp.optimize.rosen,
grad=sp.optimize.rosen_der,
hess=sp.optimize.rosen_hess)
# second type of objective
def rosen2(x):
return sp.optimize.rosen(x), sp.optimize.rosen_der(x), sp.optimize.rosen_hess(x)
objective2 = pypesto.Objective(fun=rosen2, grad=True, hess=True)
dim_full = 10
lb = -2 * np.ones((dim_full, 1))
ub = 2 * np.ones((dim_full, 1))
problem1 = pypesto.Problem(objective=objective1, lb=lb, ub=ub)
problem2 = pypesto.Problem(objective=objective2, lb=lb, ub=ub)
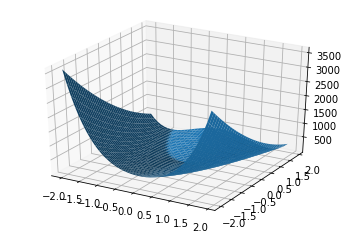
Illustration¶
[3]:
x = np.arange(-2, 2, 0.1)
y = np.arange(-2, 2, 0.1)
x, y = np.meshgrid(x, y)
z = np.zeros_like(x)
for j in range(0, x.shape[0]):
for k in range(0, x.shape[1]):
z[j,k] = objective1([x[j,k], y[j,k]], (0,))
fig = plt.figure()
ax = plt.axes(projection='3d')
ax.plot_surface(X=x, Y=y, Z=z)
[3]:
<mpl_toolkits.mplot3d.art3d.Poly3DCollection at 0x121e42908>
Run optimization¶
[4]:
optimizer = pypesto.ScipyOptimizer()
n_starts = 20
result1 = pypesto.minimize(problem=problem1, optimizer=optimizer, n_starts=n_starts)
result2 = pypesto.minimize(problem=problem2, optimizer=optimizer, n_starts=n_starts)
Compute profiles¶
The profiling routine needs a problem, a results object and an optimizer.
Moreover it accepts an index of integer (profile_index), whether or not a profile should be computed.
Finally, an integer (result_index) can be passed, in order to specify the local optimum, from which profiling should be started.
[5]:
result1 = pypesto.parameter_profile(
problem=problem1,
result=result1,
optimizer=optimizer,
profile_index=np.array([0, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0]),
result_index=1)
Visualize and analyze results¶
pypesto offers easy-to-use visualization routines:
[6]:
import pypesto.visualize
pypesto.visualize.waterfall(result1)
pypesto.visualize.parameters(result1)
# specify the parameters, for which profiles should be computed
pypesto.visualize.profiles(result1, profile_indices = [1,2,5])
[6]:
[<matplotlib.axes._subplots.AxesSubplot at 0x124363d68>,
<matplotlib.axes._subplots.AxesSubplot at 0x1245114a8>,
<matplotlib.axes._subplots.AxesSubplot at 0x1245230f0>]
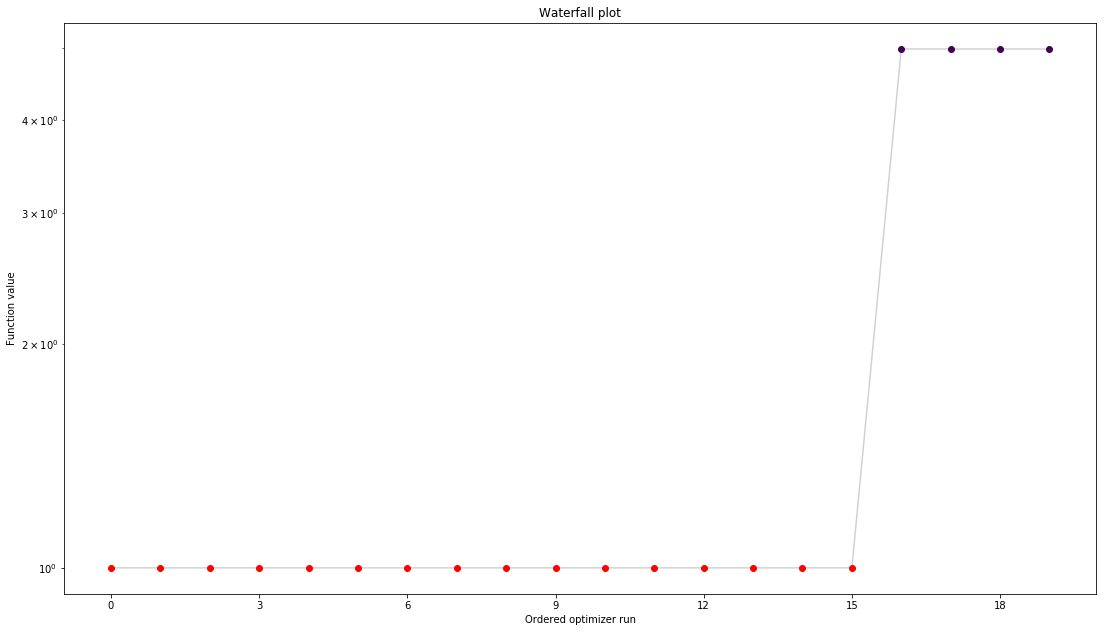
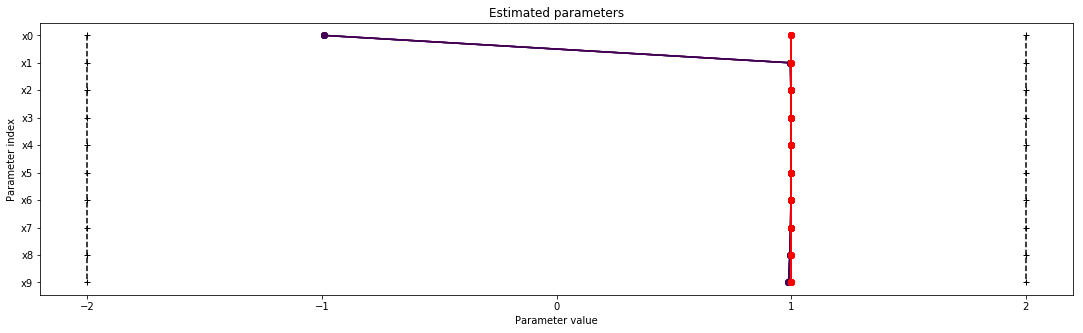
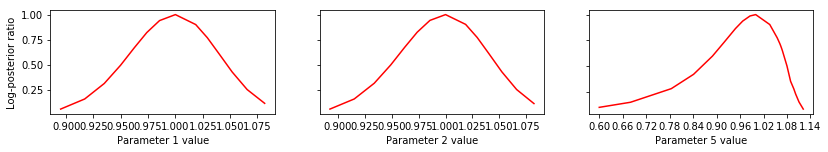
If the result needs to be examined in more detail, it can easily be exported as a pandas.DataFrame:
[7]:
result1.optimize_result.as_dataframe(['fval', 'n_fval', 'n_grad', 'n_hess', 'n_res', 'n_sres', 'time'])
[7]:
| fval | n_fval | n_grad | n_hess | n_res | n_sres | time | |
|---|---|---|---|---|---|---|---|
| 0 | 3.864669e-12 | 89 | 89 | 0 | 0 | 0 | 0.016705 |
| 1 | 5.391376e-12 | 83 | 83 | 0 | 0 | 0 | 0.015477 |
| 2 | 6.878944e-12 | 88 | 88 | 0 | 0 | 0 | 0.119707 |
| 3 | 7.215911e-12 | 91 | 91 | 0 | 0 | 0 | 0.037070 |
| 4 | 9.545020e-12 | 69 | 69 | 0 | 0 | 0 | 0.022376 |
| 5 | 1.244748e-11 | 83 | 83 | 0 | 0 | 0 | 0.022337 |
| 6 | 1.263889e-11 | 88 | 88 | 0 | 0 | 0 | 0.018748 |
| 7 | 1.562623e-11 | 86 | 86 | 0 | 0 | 0 | 0.022854 |
| 8 | 4.085166e-11 | 73 | 73 | 0 | 0 | 0 | 0.011622 |
| 9 | 7.246517e-11 | 79 | 79 | 0 | 0 | 0 | 0.026188 |
| 10 | 8.612691e-11 | 67 | 67 | 0 | 0 | 0 | 0.012368 |
| 11 | 1.823874e-10 | 76 | 76 | 0 | 0 | 0 | 0.013582 |
| 12 | 1.861598e-10 | 78 | 78 | 0 | 0 | 0 | 0.046079 |
| 13 | 1.922645e-10 | 90 | 90 | 0 | 0 | 0 | 0.025491 |
| 14 | 2.231378e-10 | 89 | 89 | 0 | 0 | 0 | 0.021747 |
| 15 | 2.499322e-10 | 73 | 73 | 0 | 0 | 0 | 0.042578 |
| 16 | 3.054340e-10 | 81 | 81 | 0 | 0 | 0 | 0.018495 |
| 17 | 3.370285e-10 | 76 | 76 | 0 | 0 | 0 | 0.017206 |
| 18 | 1.678268e-09 | 75 | 75 | 0 | 0 | 0 | 0.014224 |
| 19 | 3.986579e+00 | 71 | 71 | 0 | 0 | 0 | 0.043969 |
[ ]: