Model import using the Petab format¶
[1]:
import pypesto
import amici
import petab
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Download benchmark models - Note: 200MB :(
!git clone --depth 1 https://github.com/LoosC/Benchmark-Models.git tmp/benchmark-models || (cd tmp/benchmark-models && git pull)
fatal: destination path 'tmp/benchmark-models' already exists and is not an empty directory.
Already up to date.
Manage PETAB model¶
[2]:
folder_base = "tmp/benchmark-models/hackathon_contributions_new_data_format/"
model_name = "Zheng_PNAS2012"
model_name = "Boehm_JProteomeRes2014"
petab_problem = petab.Problem.from_folder(folder_base + model_name)
# print(petab.lint.check_measurement_df(manager.measurement_df))
petab_problem.get_optimization_to_simulation_parameter_mapping()
[2]:
[['Epo_degradation_BaF3',
'k_exp_hetero',
'k_exp_homo',
'k_imp_hetero',
'k_imp_homo',
'k_phos',
'ratio',
'sd_pSTAT5A_rel',
'sd_pSTAT5B_rel',
'sd_rSTAT5A_rel',
'specC17']]
Import model to AMICI¶
[3]:
importer = pypesto.PetabImporter(petab_problem)
model = importer.create_model()
print(model.getParameterScale())
print("Model parameters:", list(model.getParameterIds()), '\n')
print("Model const parameters:", list(model.getFixedParameterIds()), '\n')
print("Model outputs: ", list(model.getObservableIds()), '\n')
print("Model states: ", list(model.getStateIds()), '\n')
<amici.amici.ParameterScalingVector; proxy of <Swig Object of type 'std::vector< enum amici::ParameterScaling,std::allocator< enum amici::ParameterScaling > > *' at 0x12000da20> >
Model parameters: ['Epo_degradation_BaF3', 'k_exp_hetero', 'k_exp_homo', 'k_imp_hetero', 'k_imp_homo', 'k_phos', 'ratio', 'noiseParameter1_pSTAT5A_rel', 'noiseParameter1_pSTAT5B_rel', 'noiseParameter1_rSTAT5A_rel', 'specC17']
Model const parameters: []
Model outputs: ['observable_pSTAT5A_rel', 'observable_pSTAT5B_rel', 'observable_rSTAT5A_rel']
Model states: ['STAT5A', 'STAT5B', 'pApB', 'pApA', 'pBpB', 'nucpApA', 'nucpApB', 'nucpBpB']
Create objective function¶
[4]:
obj = importer.create_objective()
#print(amici.getDataObservablesAsDataFrame(obj.amici_model, edatas))
#print(edatas[0].fixedParametersPreequilibration)
#print(obj.dim, obj.x_names, len(obj.x_ids), obj.opt_to_sim_par_mapping)
#print(edatas[0].fixedParametersPreequilibration, edatas[0].fixedParameters)
print("Nominal parameter values:\n", petab_problem.x_nominal)
obj(petab_problem.x_nominal)
Nominal parameter values:
[-1.5689175884, -4.9997048936, -2.2096987817, -1.7860065475, 4.9901140088, 4.1977354885, 0.693, 0.5857552706, 0.8189828192, 0.49868440399999997, 0.107]
[4]:
138.22199805132536
Run optimization¶
[5]:
optimizer = pypesto.ScipyOptimizer()
problem = importer.create_problem(obj)
engine = pypesto.SingleCoreEngine()
# engine = pypesto.MultiProcessEngine()
# do the optimization
result = pypesto.minimize(problem=problem, optimizer=optimizer,
n_starts=5, engine=engine)
Visualize¶
[6]:
import pypesto.visualize
pypesto.visualize.waterfall(result)
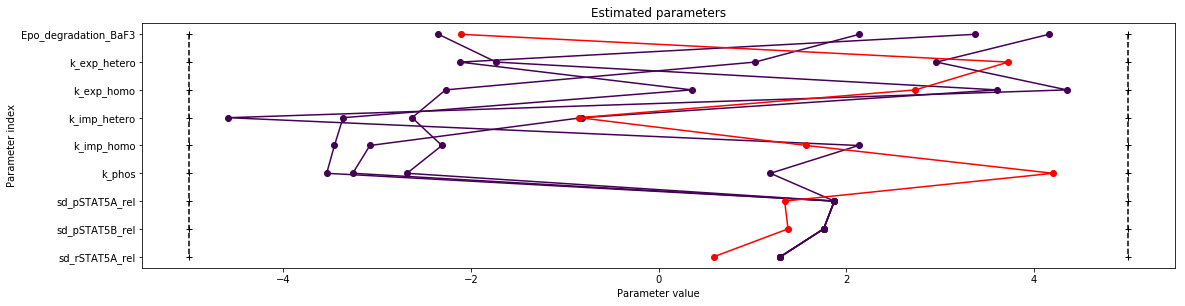
pypesto.visualize.parameters(result)
print(result.optimize_result.get_for_key('fval'))
[179.92102809791936, 249.7459637717035, 249.74599446890923, 249.74599739385891, 249.74599744289878]

[7]:
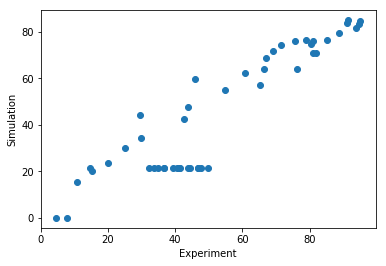
rdatas = obj(result.optimize_result.get_for_key('x')[0], return_dict=True)['rdatas']
df = importer.rdatas_to_measurement_df(rdatas)
plt.xlabel("Experiment")
plt.ylabel("Simulation")
# note: here we still assume that both files have the same order
plt.scatter(petab_problem.measurement_df['measurement'], df['measurement'])
# print(df)
[7]:
<matplotlib.collections.PathCollection at 0x1262877f0>
[ ]: