Model import using the Petab format¶
In this notebook, we illustrate using pyPESTO together with PEtab and AMICI. We employ models from the benchmark collection, which we first download:
[1]:
import pypesto
import amici
import petab
import os
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Download benchmark models - Note: 200MB :(
!git clone --depth 1 https://github.com/Benchmarking-Initiative/Benchmark-Models-PEtab.git tmp/benchmark-models || (cd tmp/benchmark-models && git pull)
folder_base = "tmp/benchmark-models/Benchmark-Models/"
fatal: destination path 'tmp/benchmark-models' already exists and is not an empty directory.
Already up to date.
Manage PEtab model¶
A PEtab problem comprises all the information on the model, the data and the parameters to perform parameter estimation:
[2]:
# a collection of models that can be simulated
#model_name = "Zheng_PNAS2012"
model_name = "Boehm_JProteomeRes2014"
#model_name = "Fujita_SciSignal2010"
#model_name = "Sneyd_PNAS2002"
#model_name = "Borghans_BiophysChem1997"
#model_name = "Elowitz_Nature2000"
#model_name = "Crauste_CellSystems2017"
#model_name = "Lucarelli_CellSystems2018"
#model_name = "Schwen_PONE2014"
#model_name = "Blasi_CellSystems2016"
# the yaml configuration file links to all needed files
yaml_config = os.path.join(folder_base, model_name, model_name + '.yaml')
# create a petab problem
petab_problem = petab.Problem.from_yaml(yaml_config)
Import model to AMICI¶
The model must be imported to AMICI:
[3]:
importer = pypesto.PetabImporter(petab_problem)
model = importer.create_model()
print("Model parameters:", list(model.getParameterIds()), '\n')
print("Model const parameters:", list(model.getFixedParameterIds()), '\n')
print("Model outputs: ", list(model.getObservableIds()), '\n')
print("Model states: ", list(model.getStateIds()), '\n')
Model parameters: ['Epo_degradation_BaF3', 'k_exp_hetero', 'k_exp_homo', 'k_imp_hetero', 'k_imp_homo', 'k_phos', 'ratio', 'specC17', 'noiseParameter1_pSTAT5A_rel', 'noiseParameter1_pSTAT5B_rel', 'noiseParameter1_rSTAT5A_rel']
Model const parameters: []
Model outputs: ['pSTAT5A_rel', 'pSTAT5B_rel', 'rSTAT5A_rel']
Model states: ['STAT5A', 'STAT5B', 'pApB', 'pApA', 'pBpB', 'nucpApA', 'nucpApB', 'nucpBpB']
Create objective function¶
[4]:
import libsbml
converter_config = libsbml.SBMLLocalParameterConverter()\
.getDefaultProperties()
petab_problem.sbml_document.convert(converter_config)
obj = importer.create_objective()
# for some models, hyperparamters need to be adjusted
#obj.amici_solver.setMaxSteps(10000)
#obj.amici_solver.setRelativeTolerance(1e-7)
#obj.amici_solver.setAbsoluteTolerance(1e-7)
ret = obj(petab_problem.x_nominal_scaled, sensi_orders=(0,1), return_dict=True)
print(ret)
{'fval': 138.22199677513575, 'grad': array([ 2.20386015e-02, 5.53227506e-02, 5.78886452e-03, 5.40656415e-03,
-4.51595809e-05, 7.91163446e-03, 0.00000000e+00, 1.07840959e-02,
2.40378735e-02, 1.91919657e-02, 0.00000000e+00]), 'hess': array([[-2.11105595e+03, -5.89390039e-01, -1.07159910e+02,
-2.81393973e+03, -8.94333861e-06, 7.86055092e+02,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-5.89390039e-01, -1.91513744e-03, 1.72774945e-01,
-7.12558479e-01, 3.69774927e-08, 3.20531692e-01,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-1.07159910e+02, 1.72774945e-01, -6.99839693e+01,
-1.61497679e+02, -7.16323554e-06, 8.83572656e+01,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-2.81393973e+03, -7.12558479e-01, -1.61497679e+02,
-3.76058352e+03, -8.40044683e-06, 1.04136909e+03,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-8.94333861e-06, 3.69774927e-08, -7.16323554e-06,
-8.40044683e-06, -2.86438192e-10, 2.24927732e-04,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 7.86055092e+02, 3.20531692e-01, 8.83572656e+01,
1.04136909e+03, 2.24927732e-04, -9.29902113e+02,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00]]), 'res': array([], dtype=float64), 'sres': array([], shape=(0, 11), dtype=float64), 'rdatas': [<amici.numpy.ReturnDataView object at 0x7f247cedec10>]}
For debugging: There is an alternative way of computing the function, in amici directly.
[5]:
import libsbml
converter_config = libsbml.SBMLLocalParameterConverter()\
.getDefaultProperties()
petab_problem.sbml_document.convert(converter_config)
obj2 = importer.create_objective()
obj2.use_amici_petab_simulate = True
# for some models, hyperparamters need to be adjusted
#obj2.amici_solver.setMaxSteps(int(1e8))
#obj2.amici_solver.setRelativeTolerance(1e-3)
#obj2.amici_solver.setAbsoluteTolerance(1e-3)
ret2 = obj2(petab_problem.x_nominal_scaled, sensi_orders=(0,1), return_dict=True)
print(ret2)
{'fval': 138.22199677513575, 'grad': array([ 2.20386015e-02, 5.53227506e-02, 5.78886452e-03, 5.40656415e-03,
-4.51595809e-05, 7.91163446e-03, 0.00000000e+00, 1.07840959e-02,
2.40378735e-02, 1.91919657e-02, 0.00000000e+00]), 'hess': array([[-2.11105595e+03, -5.89390039e-01, -1.07159910e+02,
-2.81393973e+03, -8.94333861e-06, 7.86055092e+02,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-5.89390039e-01, -1.91513744e-03, 1.72774945e-01,
-7.12558479e-01, 3.69774927e-08, 3.20531692e-01,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-1.07159910e+02, 1.72774945e-01, -6.99839693e+01,
-1.61497679e+02, -7.16323554e-06, 8.83572656e+01,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-2.81393973e+03, -7.12558479e-01, -1.61497679e+02,
-3.76058352e+03, -8.40044683e-06, 1.04136909e+03,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[-8.94333861e-06, 3.69774927e-08, -7.16323554e-06,
-8.40044683e-06, -2.86438192e-10, 2.24927732e-04,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 7.86055092e+02, 3.20531692e-01, 8.83572656e+01,
1.04136909e+03, 2.24927732e-04, -9.29902113e+02,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00],
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00]]), 'res': array([], dtype=float64), 'sres': array([], shape=(0, 11), dtype=float64), 'rdatas': [<amici.numpy.ReturnDataView object at 0x7f247cba3290>]}
A finite difference check whether the computed gradient is accurate:
[6]:
problem = importer.create_problem(obj)
objective = problem.objective
ret = objective(petab_problem.x_nominal_free_scaled, sensi_orders=(0,1))
print(ret)
(138.22199677513575, array([ 2.20386015e-02, 5.53227506e-02, 5.78886452e-03, 5.40656415e-03,
-4.51595809e-05, 7.91163446e-03, 1.07840959e-02, 2.40378735e-02,
1.91919657e-02]))
[7]:
eps = 1e-4
def fd(x):
grad = np.zeros_like(x)
j = 0
for i, xi in enumerate(x):
mask = np.zeros_like(x)
mask[i] += eps
valinc, _ = objective(x+mask, sensi_orders=(0,1))
valdec, _ = objective(x-mask, sensi_orders=(0,1))
grad[j] = (valinc - valdec) / (2*eps)
j += 1
return grad
fdval = fd(petab_problem.x_nominal_free_scaled)
print("fd: ", fdval)
print("l2 difference: ", np.linalg.norm(ret[1] - fdval))
fd: [0.02493368 0.05309659 0.00530587 0.01291083 0.00587754 0.01473653
0.01078279 0.02403657 0.01919066]
l2 difference: 0.012310244824532144
Run optimization¶
[8]:
print(problem.x_fixed_indices, problem.x_free_indices)
[6, 10] [0, 1, 2, 3, 4, 5, 7, 8, 9]
[9]:
optimizer = pypesto.ScipyOptimizer()
engine = pypesto.SingleCoreEngine()
engine = pypesto.MultiProcessEngine()
# do the optimization
result = pypesto.minimize(problem=problem, optimizer=optimizer,
n_starts=10, engine=engine)
Engine set up to use up to 4 processes in total. The number was automatically determined and might not be appropriate on some systems.
Visualize¶
[10]:
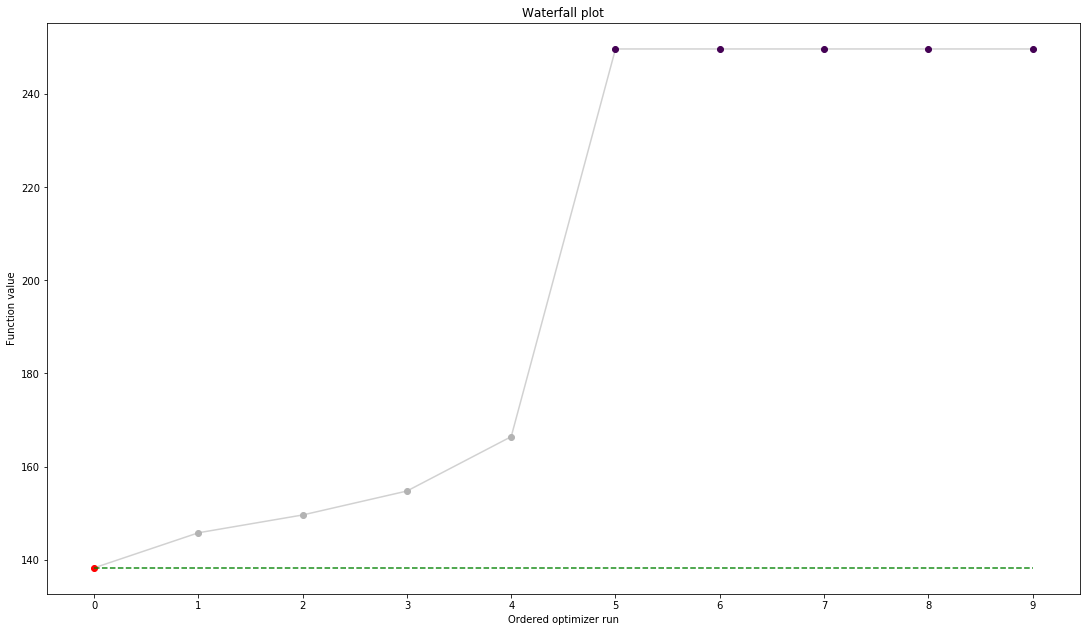
result.optimize_result.get_for_key('fval')
[10]:
[138.22197383818877,
145.75940996955092,
149.58861509227683,
154.73313632726362,
166.42016015420128,
249.7445771549656,
249.74598291093008,
249.74598526715747,
249.74599456039678,
249.74599744358338]
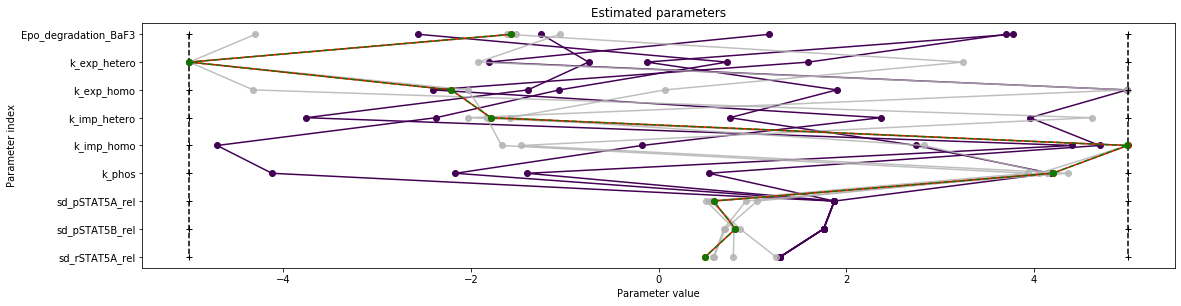
[11]:
import pypesto.visualize
ref = pypesto.visualize.create_references(x=petab_problem.x_nominal_scaled, fval=obj(petab_problem.x_nominal_scaled))
pypesto.visualize.waterfall(result, reference=ref, scale_y='lin')
pypesto.visualize.parameters(result, reference=ref)
[11]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f247bd8bb90>